MDSVDNV: predicting microbe-drug associations by singular value decomposition and Node2vec
- PMID: 38260900
- PMCID: PMC10800927
- DOI: 10.3389/fmicb.2023.1303585
MDSVDNV: predicting microbe-drug associations by singular value decomposition and Node2vec
Abstract
Introduction: Recent researches have demonstrated that microbes are crucial for the growth and development of the human body, the movement of nutrients, and human health. Diseases may arise as a result of disruptions and imbalances in the microbiome. The pathological investigation of associated diseases and the advancement of clinical medicine can both benefit from the identification of drug-associated microbes.
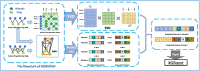
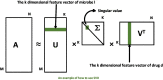
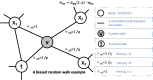
Methods: In this article, we proposed a new prediction model called MDSVDNV to infer potential microbe-drug associations, in which the Node2vec network embedding approach and the singular value decomposition (SVD) matrix decomposition method were first adopted to produce linear and non-linear representations of microbe interactions.
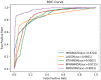
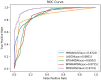
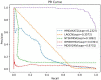
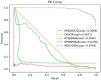
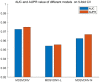
Results and discussion: Compared with state-of-the-art competitive methods, intensive experimental results demonstrated that MDSVDNV could achieve the best AUC value of 98.51% under a 5-fold CV, which indicated that MDSVDNV outperformed existing competing models and may be an effective method for discovering latent microbe-drug associations in the future.
Keywords: Node2vec; XGBoost classifier; computational model; microbe–drug association prediction; singular value decomposition.
Copyright © 2024 Tan, Zhang, Liu, Chen, Yang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
SVDNVLDA: predicting lncRNA-disease associations by Singular Value Decomposition and node2vec.BMC Bioinformatics. 2021 Nov 2;22(1):538. doi: 10.1186/s12859-021-04457-1. BMC Bioinformatics. 2021. PMID: 34727886 Free PMC article.
-
LCASPMDA: a computational model for predicting potential microbe-drug associations based on learnable graph convolutional attention networks and self-paced iterative sampling ensemble.Front Microbiol. 2024 May 23;15:1366272. doi: 10.3389/fmicb.2024.1366272. eCollection 2024. Front Microbiol. 2024. PMID: 38846568 Free PMC article.
-
GSAMDA: a computational model for predicting potential microbe-drug associations based on graph attention network and sparse autoencoder.BMC Bioinformatics. 2022 Nov 18;23(1):492. doi: 10.1186/s12859-022-05053-7. BMC Bioinformatics. 2022. PMID: 36401174 Free PMC article.
-
Prediction of drug-disease associations based on ensemble meta paths and singular value decomposition.BMC Bioinformatics. 2019 Mar 29;20(Suppl 3):134. doi: 10.1186/s12859-019-2644-5. BMC Bioinformatics. 2019. PMID: 30925858 Free PMC article.
-
MCHMDA:Predicting Microbe-Disease Associations Based on Similarities and Low-Rank Matrix Completion.IEEE/ACM Trans Comput Biol Bioinform. 2021 Mar-Apr;18(2):611-620. doi: 10.1109/TCBB.2019.2926716. Epub 2021 Apr 12. IEEE/ACM Trans Comput Biol Bioinform. 2021. PMID: 31295117
Cited by
-
GLNNMDA: a multimodal prediction model for microbe-drug associations based on global and local features.Sci Rep. 2024 Sep 6;14(1):20847. doi: 10.1038/s41598-024-71837-x. Sci Rep. 2024. PMID: 39242712 Free PMC article.
-
GiGs: graph-based integrated Gaussian kernel similarity for virus-drug association prediction.Brief Bioinform. 2025 Mar 4;26(2):bbaf117. doi: 10.1093/bib/bbaf117. Brief Bioinform. 2025. PMID: 40112339 Free PMC article.
References
LinkOut - more resources
Full Text Sources

