Cis-regulatory modes of Ultrabithorax inactivation in butterfly forewings
- PMID: 38261357
- PMCID: PMC10945631
- DOI: 10.7554/eLife.90846
Cis-regulatory modes of Ultrabithorax inactivation in butterfly forewings
Abstract
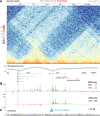
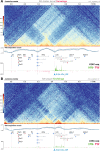
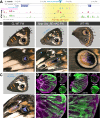
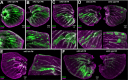
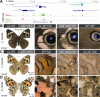
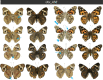
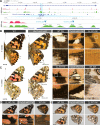
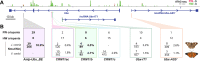
Hox gene clusters encode transcription factors that drive regional specialization during animal development: for example the Hox factor Ubx is expressed in the insect metathoracic (T3) wing appendages and differentiates them from T2 mesothoracic identities. Hox transcriptional regulation requires silencing activities that prevent spurious activation and regulatory crosstalks in the wrong tissues, but this has seldom been studied in insects other than Drosophila, which shows a derived Hox dislocation into two genomic clusters that disjoined Antennapedia (Antp) and Ultrabithorax (Ubx). Here, we investigated how Ubx is restricted to the hindwing in butterflies, amidst a contiguous Hox cluster. By analysing Hi-C and ATAC-seq data in the butterfly Junonia coenia, we show that a Topologically Associated Domain (TAD) maintains a hindwing-enriched profile of chromatin opening around Ubx. This TAD is bordered by a Boundary Element (BE) that separates it from a region of joined wing activity around the Antp locus. CRISPR mutational perturbation of this BE releases ectopic Ubx expression in forewings, inducing homeotic clones with hindwing identities. Further mutational interrogation of two non-coding RNA encoding regions and one putative cis-regulatory module within the Ubx TAD cause rare homeotic transformations in both directions, indicating the presence of both activating and repressing chromatin features. We also describe a series of spontaneous forewing homeotic phenotypes obtained in Heliconius butterflies, and discuss their possible mutational basis. By leveraging the extensive wing specialization found in butterflies, our initial exploration of Ubx regulation demonstrates the existence of silencing and insulating sequences that prevent its spurious expression in forewings.
Keywords: Hox; boundary element; butterflies; evo-devo; evolutionary biology; genetics; genomics.
© 2023, Tendolkar et al.
Conflict of interest statement
AT, AM, LL, JH, KV, LG, AM No competing interests declared
Figures













Update of
- doi: 10.1101/2023.08.18.553910
- doi: 10.7554/eLife.90846.1
- doi: 10.7554/eLife.90846.2
References
-
- Anania C, Acemel RD, Jedamzick J, Bolondi A, Cova G, Brieske N, Kühn R, Wittler L, Real FM, Lupiáñez DG. In vivo dissection of a clustered-CTCF domain boundary reveals developmental principles of regulatory insulation. Nature Genetics. 2022;54:1026–1036. doi: 10.1038/s41588-022-01117-9. - DOI - PMC - PubMed
-
- Armstrong J, Hickey G, Diekhans M, Fiddes IT, Novak AM, Deran A, Fang Q, Xie D, Feng S, Stiller J, Genereux D, Johnson J, Marinescu VD, Alföldi J, Harris RS, Lindblad-Toh K, Haussler D, Karlsson E, Jarvis ED, Zhang G, Paten B. Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature. 2020;587:246–251. doi: 10.1038/s41586-020-2871-y. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

