Exploring Immunome and Microbiome Interplay in Reproductive Health: Current Knowledge, Challenges, and Novel Diagnostic Tools
- PMID: 38262441
- PMCID: PMC10846929
- DOI: 10.1055/s-0043-1778017
Exploring Immunome and Microbiome Interplay in Reproductive Health: Current Knowledge, Challenges, and Novel Diagnostic Tools
Abstract
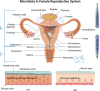
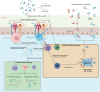
The dynamic interplay between the immunome and microbiome in reproductive health is a complex and rapidly advancing research field, holding tremendously vast possibilities for the development of reproductive medicine. This immunome-microbiome relationship influences the innate and adaptive immune responses, thereby affecting the onset and progression of reproductive disorders. However, the mechanisms governing these interactions remain elusive and require innovative approaches to gather more understanding. This comprehensive review examines the current knowledge on reproductive microbiomes across various parts of female reproductive tract, with special consideration of bidirectional interactions between microbiomes and the immune system. Additionally, it explores innate and adaptive immunity, focusing on immunoglobulin (Ig) A and IgM antibodies, their regulation, self-antigen tolerance mechanisms, and their roles in immune homeostasis. This review also highlights ongoing technological innovations in microbiota research, emphasizing the need for standardized detection and analysis methods. For instance, we evaluate the clinical utility of innovative technologies such as Phage ImmunoPrecipitation Sequencing (PhIP-Seq) and Microbial Flow Cytometry coupled to Next-Generation Sequencing (mFLOW-Seq). Despite ongoing advancements, we emphasize the need for further exploration in this field, as a deeper understanding of immunome-microbiome interactions holds promise for innovative diagnostic and therapeutic strategies for reproductive health, like infertility treatment and management of pregnancy.
The Author(s). This is an open access article published by Thieme under the terms of the Creative Commons Attribution-NonDerivative-NonCommercial License, permitting copying and reproduction so long as the original work is given appropriate credit. Contents may not be used for commercial purposes, or adapted, remixed, transformed or built upon. (https://creativecommons.org/licenses/by-nc-nd/4.0/).
Conflict of interest statement
All authors declare that they have no competing interests.
Figures



Similar articles
-
Deciphering the role of female reproductive tract microbiome in reproductive health: a review.Front Cell Infect Microbiol. 2024 Mar 18;14:1351540. doi: 10.3389/fcimb.2024.1351540. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38562966 Free PMC article. Review.
-
The Interplay Between Reproductive Tract Microbiota and Immunological System in Human Reproduction.Front Immunol. 2020 Mar 16;11:378. doi: 10.3389/fimmu.2020.00378. eCollection 2020. Front Immunol. 2020. PMID: 32231664 Free PMC article. Review.
-
The impact of the female genital tract microbiome in women health and reproduction: a review.J Assist Reprod Genet. 2021 Oct;38(10):2519-2541. doi: 10.1007/s10815-021-02247-5. Epub 2021 Jun 10. J Assist Reprod Genet. 2021. PMID: 34110573 Free PMC article. Review.
-
Identification and evaluation of the microbiome in the female and male reproductive tracts.Hum Reprod Update. 2019 May 1;25(3):298-325. doi: 10.1093/humupd/dmy048. Hum Reprod Update. 2019. PMID: 30938752 Review.
-
Microbiome in Female Reproductive Health: Implications for Fertility and Assisted Reproductive Technologies.Genomics Proteomics Bioinformatics. 2024 May 9;22(1):qzad005. doi: 10.1093/gpbjnl/qzad005. Genomics Proteomics Bioinformatics. 2024. PMID: 38862423 Free PMC article. Review.
Cited by
-
Plasma Cells as the Key Players of IVF Failure? Unlocking the Enigma of Infertility and In Vitro Fertilization Failure in the Light of Uterine Inflammation.Int J Mol Sci. 2024 Dec 5;25(23):13083. doi: 10.3390/ijms252313083. Int J Mol Sci. 2024. PMID: 39684794 Free PMC article. Review.
-
The evolving facets of vaginal microbiota transplantation: reinvigorating the unexplored frontier amid complex challenges.Arch Microbiol. 2024 Jun 15;206(7):306. doi: 10.1007/s00203-024-04024-1. Arch Microbiol. 2024. PMID: 38878076 Review.
-
Gut Microbiome Dysbiosis and Its Impact on Reproductive Health: Mechanisms and Clinical Applications.Metabolites. 2025 Jun 11;15(6):390. doi: 10.3390/metabo15060390. Metabolites. 2025. PMID: 40559414 Free PMC article. Review.
References
-
- Wang B, Yao M, Lv L, Ling Z, Li L. The human microbiota in health and disease. Engineering. 2017;3(01):71–82.
-
- Brüls T, Weissenbach J.The human metagenome: our other genome? Hum Mol Genet 201120(R2):R142–R148. - PubMed
-
- Donders G GG, Bellen G, Ruban K S. Abnormal vaginal microbiome is associated with severity of localized provoked vulvodynia. Role of aerobic vaginitis and Candida in the pathogenesis of vulvodynia. Eur J Clin Microbiol Infect Dis. 2018;37(09):1679–1685. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

