Efficacy of MRI data harmonization in the age of machine learning: a multicenter study across 36 datasets
- PMID: 38263181
- PMCID: PMC10805868
- DOI: 10.1038/s41597-023-02421-7
Efficacy of MRI data harmonization in the age of machine learning: a multicenter study across 36 datasets
Abstract
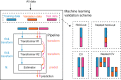
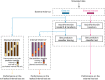
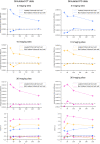
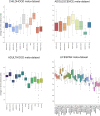
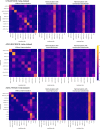
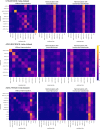
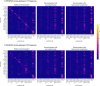
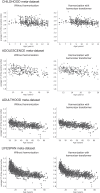
Pooling publicly-available MRI data from multiple sites allows to assemble extensive groups of subjects, increase statistical power, and promote data reuse with machine learning techniques. The harmonization of multicenter data is necessary to reduce the confounding effect associated with non-biological sources of variability in the data. However, when applied to the entire dataset before machine learning, the harmonization leads to data leakage, because information outside the training set may affect model building, and potentially falsely overestimate performance. We propose a 1) measurement of the efficacy of data harmonization; 2) harmonizer transformer, i.e., an implementation of the ComBat harmonization allowing its encapsulation among the preprocessing steps of a machine learning pipeline, avoiding data leakage by design. We tested these tools using brain T1-weighted MRI data from 1740 healthy subjects acquired at 36 sites. After harmonization, the site effect was removed or reduced, and we showed the data leakage effect in predicting individual age from MRI data, highlighting that introducing the harmonizer transformer into a machine learning pipeline allows for avoiding data leakage by design.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Performance comparison of modified ComBat for harmonization of radiomic features for multicenter studies.Sci Rep. 2020 Jun 24;10(1):10248. doi: 10.1038/s41598-020-66110-w. Sci Rep. 2020. PMID: 32581221 Free PMC article.
-
Effect of data harmonization of multicentric dataset in ASD/TD classification.Brain Inform. 2023 Nov 25;10(1):32. doi: 10.1186/s40708-023-00210-x. Brain Inform. 2023. PMID: 38006422 Free PMC article.
-
Comparison of traveling-subject and ComBat harmonization methods for assessing structural brain characteristics.Hum Brain Mapp. 2021 Nov;42(16):5278-5287. doi: 10.1002/hbm.25615. Epub 2021 Aug 17. Hum Brain Mapp. 2021. PMID: 34402132 Free PMC article.
-
Deep Learning in Large and Multi-Site Structural Brain MR Imaging Datasets.Front Neuroinform. 2022 Jan 20;15:805669. doi: 10.3389/fninf.2021.805669. eCollection 2021. Front Neuroinform. 2022. PMID: 35126080 Free PMC article. Review.
-
Convolutional neural networks for classification of Alzheimer's disease: Overview and reproducible evaluation.Med Image Anal. 2020 Jul;63:101694. doi: 10.1016/j.media.2020.101694. Epub 2020 May 1. Med Image Anal. 2020. PMID: 32417716
Cited by
-
Deep Learning for MRI Segmentation and Molecular Subtyping in Glioblastoma: Critical Aspects from an Emerging Field.Biomedicines. 2024 Aug 16;12(8):1878. doi: 10.3390/biomedicines12081878. Biomedicines. 2024. PMID: 39200342 Free PMC article. Review.
-
A critical assessment of artificial intelligence in magnetic resonance imaging of cancer.Npj Imaging. 2025;3(1):15. doi: 10.1038/s44303-025-00076-0. Epub 2025 Apr 9. Npj Imaging. 2025. PMID: 40226507 Free PMC article. Review.
-
Editorial: Methods and application in fractal analysis of neuroimaging data.Front Hum Neurosci. 2024 Jul 10;18:1453284. doi: 10.3389/fnhum.2024.1453284. eCollection 2024. Front Hum Neurosci. 2024. PMID: 39050380 Free PMC article. No abstract available.
-
Lifespan reference curves for harmonizing multi-site regional brain white matter metrics from diffusion MRI.bioRxiv [Preprint]. 2024 Mar 1:2024.02.22.581646. doi: 10.1101/2024.02.22.581646. bioRxiv. 2024. Update in: Sci Data. 2025 May 6;12(1):748. doi: 10.1038/s41597-025-05028-2. PMID: 38463962 Free PMC article. Updated. Preprint.
-
Superpixel-ComBat modeling: A joint approach for harmonization and characterization of inter-scanner variability in T1-weighted images.Imaging Neurosci (Camb). 2024 Oct 3;2:imag-2-00306. doi: 10.1162/imag_a_00306. eCollection 2024. Imaging Neurosci (Camb). 2024. PMID: 40800451 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

