Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM
- PMID: 38263358
- PMCID: PMC10805800
- DOI: 10.1038/s41421-023-00639-8
Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM
Erratum in
-
Author Correction: Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM.Cell Discov. 2024 Feb 26;10(1):23. doi: 10.1038/s41421-024-00660-5. Cell Discov. 2024. PMID: 38409077 Free PMC article. No abstract available.
Abstract
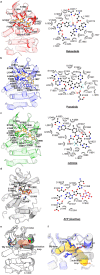
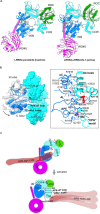
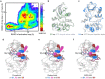
LRRK2 is one of the most promising drug targets for Parkinson's disease. Though type I kinase inhibitors of LRRK2 are under clinical trials, alternative strategies like type II inhibitors are being actively pursued due to the potential undesired effects of type I inhibitors. Currently, a robust method for LRRK2-inhibitor structure determination to guide structure-based drug discovery is lacking, and inhibition mechanisms of available compounds are also unclear. Here we present near-atomic-resolution structures of LRRK2 with type I (LRRK2-IN-1 and GNE-7915) and type II (rebastinib, ponatinib, and GZD-824) inhibitors, uncovering the structural basis of LRRK2 inhibition and conformational plasticity of the kinase domain with molecular dynamics (MD) simulations. Type I and II inhibitors bind to LRRK2 in active-like and inactive conformations, so LRRK2-inhibitor complexes further reveal general structural features associated with LRRK2 activation. Our study provides atomic details of LRRK2-inhibitor interactions and a framework for understanding LRRK2 activation and for rational drug design.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Alexander Boecker C. The role of LRRK2 in intracellular organelle dynamics. J. Mol. Biol. 2023;435:167998. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

