Effect of structural variation in the promoter region of RsMYB1.1 on the skin color of radish taproot
- PMID: 38264015
- PMCID: PMC10804855
- DOI: 10.3389/fpls.2023.1327009
Effect of structural variation in the promoter region of RsMYB1.1 on the skin color of radish taproot
Abstract
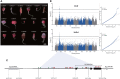
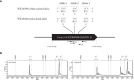
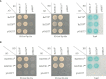
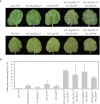
Accumulation of anthocyanins in the taproot of radish is an agronomic trait beneficial for human health. Several genetic loci are related to a red skin or flesh color of radish, however, the functional divergence of candidate genes between non-red and red radishes has not been investigated. Here, we report that a novel genetic locus on the R2 chromosome, where RsMYB1.1 is located, is associated with the red color of the skin of radish taproot. A genome-wide association study (GWAS) of 66 non-red-skinned (nR) and 34 red-skinned (R) radish accessions identified three nonsynonymous single nucleotide polymorphisms (SNPs) in the third exon of RsMYB1.1. Although the genotypes of SNP loci differed between the nR and R radishes, no functional difference in the RsMYB1.1 proteins of nR and R radishes in their physical interaction with RsTT8 was detected by yeast-two hybrid assay or in anthocyanin accumulation in tobacco and radish leaves coexpressing RsMYB1.1 and RsTT8. By contrast, insertion- or deletion-based GWAS revealed that one large AT-rich low-complexity sequence of 1.3-2 kb was inserted in the promoter region of RsMYB1.1 in the nR radishes (RsMYB1.1nR), whereas the R radishes had no such insertion; this represents a presence/absence variation (PAV). This insertion sequence (RsIS) was radish specific and distributed among the nine chromosomes of Raphanus genomes. Despite the extremely low transcription level of RsMYB1.1nR in the nR radishes, the inactive RsMYB1.1nR promoter could be functionally restored by deletion of the RsIS. The results of a transient expression assay using radish root sections suggested that the RsIS negatively regulates the expression of RsMYB1.1nR, resulting in the downregulation of anthocyanin biosynthesis genes, including RsCHS, RsDFR, and RsANS, in the nR radishes. This work provides the first evidence of the involvement of PAV in an agronomic trait of radish.
Keywords: GWAS; RsMYB1.1; anthocyanin; presence/absence variation; promoter; radish; red skin color.
Copyright © 2024 Kim, Jang, Huh, Cho, Yim, Jeong, Kim, Yu and Mun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Loss of the R2R3 MYB Transcription Factor RsMYB1 Shapes Anthocyanin Biosynthesis and Accumulation in Raphanus sativus.Int J Mol Sci. 2021 Oct 10;22(20):10927. doi: 10.3390/ijms222010927. Int J Mol Sci. 2021. PMID: 34681588 Free PMC article.
-
A Radish Basic Helix-Loop-Helix Transcription Factor, RsTT8 Acts a Positive Regulator for Anthocyanin Biosynthesis.Front Plant Sci. 2017 Nov 8;8:1917. doi: 10.3389/fpls.2017.01917. eCollection 2017. Front Plant Sci. 2017. PMID: 29167678 Free PMC article.
-
Intron-retained radish (Raphanus sativus L.) RsMYB1 transcripts found in colored-taproot lines enhance anthocyanin accumulation in transgenic Arabidopsis plants.Plant Cell Rep. 2021 Sep;40(9):1735-1749. doi: 10.1007/s00299-021-02735-z. Epub 2021 Jul 25. Plant Cell Rep. 2021. PMID: 34308490
-
Differential anthocyanin accumulation in radish taproot: importance of RsMYB1 gene structure.Plant Cell Rep. 2020 Feb;39(2):217-226. doi: 10.1007/s00299-019-02485-z. Epub 2019 Nov 14. Plant Cell Rep. 2020. PMID: 31728702
-
Transposon-induced methylation of the RsMYB1 promoter disturbs anthocyanin accumulation in red-fleshed radish.J Exp Bot. 2020 May 9;71(9):2537-2550. doi: 10.1093/jxb/eraa010. J Exp Bot. 2020. PMID: 31961436 Free PMC article.
Cited by
-
Structure and evolution of the Forsythieae genome elucidated by chromosome-level genome comparison of Abeliophyllum distichum and Forsythia ovata (Oleaceae).Commun Biol. 2025 Feb 18;8(1):254. doi: 10.1038/s42003-025-07683-y. Commun Biol. 2025. PMID: 39966682 Free PMC article.
References
-
- Arro J., Labate J. (2022). Genetic variation in a radish (Raphanus sativus L.) geodiversity collection. Genet. Resour Crop Evol. 69, 163–171. doi: 10.1007/s10722-021-01212-6 - DOI
LinkOut - more resources
Full Text Sources

