A targeted approach to enrich host-associated bacteria for metagenomic sequencing
- PMID: 38264162
- PMCID: PMC10804224
- DOI: 10.1093/femsmc/xtad021
A targeted approach to enrich host-associated bacteria for metagenomic sequencing
Abstract
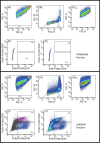
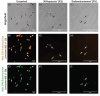
Multicellular eukaryotic organisms are hosts to communities of bacteria that reside on or inside their tissues. Often the eukaryotic members of the system contribute to high proportions of metagenomic sequencing reads, making it challenging to achieve sufficient sequencing depth to evaluate bacterial ecology. Stony corals are one such complex community; however, separation of bacterial from eukaryotic (primarily coral and algal symbiont) cells has so far not been successful. Using a combination of hybridization chain reaction fluorescence in situ hybridization and fluorescence activated cell sorting (HCR-FISH + FACS), we sorted two populations of bacteria from five genotypes of the coral Acropora loripes, targeting (i) Endozoicomonas spp, and (ii) all other bacteria. NovaSeq sequencing resulted in 67-91 M reads per sample, 55%-90% of which were identified as bacterial. Most reads were taxonomically assigned to the key coral-associated family, Endozoicomonadaceae, with Vibrionaceae also abundant. Endozoicomonadaceae were 5x more abundant in the 'Endozoicomonas' population, highlighting the success of the dual-labelling approach. This method effectively enriched coral samples for bacteria with <1% contamination from host and algal symbionts. The application of this method will allow researchers to decipher the functional potential of coral-associated bacteria. This method can also be adapted to accommodate other host-associated communities.
Keywords: FACS; FISH; coral; host-associated; hybridization chain reaction; metagenomics; symbiosis.
© The Author(s) 2023. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Targeted single-cell genomics reveals novel host adaptation strategies of the symbiotic bacteria Endozoicomonas in Acropora tenuis coral.Microbiome. 2022 Dec 12;10(1):220. doi: 10.1186/s40168-022-01395-9. Microbiome. 2022. PMID: 36503599 Free PMC article.
-
Differential aggregation patterns of Endozoicomonas within tissues of the coral Acropora loripes.ISME J. 2025 Mar 28:wraf059. doi: 10.1093/ismejo/wraf059. Online ahead of print. ISME J. 2025. PMID: 40156309
-
Microbiomes of stony and soft deep-sea corals share rare core bacteria.Microbiome. 2019 Jun 10;7(1):90. doi: 10.1186/s40168-019-0697-3. Microbiome. 2019. PMID: 31182168 Free PMC article.
-
Ecology of Endozoicomonadaceae in three coral genera across the Pacific Ocean.Nat Commun. 2023 Jun 1;14(1):3037. doi: 10.1038/s41467-023-38502-9. Nat Commun. 2023. PMID: 37264015 Free PMC article.
-
Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas.Appl Microbiol Biotechnol. 2016 Oct;100(19):8315-24. doi: 10.1007/s00253-016-7777-0. Epub 2016 Aug 24. Appl Microbiol Biotechnol. 2016. PMID: 27557714 Free PMC article. Review.
Cited by
-
Chlamydiae in corals: shared functional potential despite broad taxonomic diversity.ISME Commun. 2024 Apr 15;4(1):ycae054. doi: 10.1093/ismeco/ycae054. eCollection 2024 Jan. ISME Commun. 2024. PMID: 38707840 Free PMC article.
-
DNA from non-viable bacteria biases diversity estimates in the corals Acropora loripes and Pocillopora acuta.Environ Microbiome. 2023 Dec 8;18(1):86. doi: 10.1186/s40793-023-00541-6. Environ Microbiome. 2023. PMID: 38062479 Free PMC article.
-
Changes in bumblebee queen gut microbiotas during and after overwintering diapause.Insect Mol Biol. 2025 Feb;34(1):136-150. doi: 10.1111/imb.12957. Epub 2024 Aug 22. Insect Mol Biol. 2025. PMID: 39175129 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
