Multi-tissue transcriptome analysis to identify candidate genes associated with weight regulation in Hanwoo cattle
- PMID: 38264212
- PMCID: PMC10803631
- DOI: 10.3389/fgene.2023.1304638
Multi-tissue transcriptome analysis to identify candidate genes associated with weight regulation in Hanwoo cattle
Abstract
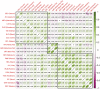
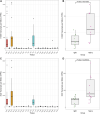
While genetic markers related to meat production traits have been identified in many other cattle breeds, research on weight in Hanwoo cattle (Korean native cattle) is still insufficient. In this study, we performed expression quantitative trait loci (eQTL) analysis and differential gene expression analysis to detect candidate genes influencing the weight characteristics of 32 castrated Hanwoo cattle across 22 tissues and, we identified variants that affect gene expression levels. In total, we identified a total of 3,298 differentially expressed genes, among which we discovered key genes such as UBD, RGS2, FASN, and SCD that have functions related to adipogenesis, body weight, obesity, and lipid metabolism. Gene-set enrichment analysis revealed that candidate genes in adipose tissue are involved in metabolic pathways linked to obesity-related traits, adipose metabolism, and lipid metabolism. Additionally, we found that decreased expression of TRIM31 contributes to weight gain which can be explained by the associated candidate cis-eQTL genotypes for TRIM31 and their effect on differential gene expression between the lower and higher weight groups. Our findings revealed candidate genes associated with the weight of Hanwoo cattle and perhaps can provide comprehensive insights into the association of weight with various tissues beyond adipose tissue and muscle, indicating the potential for expanding the focus of livestock trait research.
Keywords: Hanwoo; body weight; differentially expressed genes; eQTL analysis; transcriptome.
Copyright © 2024 Jang, Jang, Kim and Park.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Cai W., Zhang Y., Chang T., Wang Z., Zhu B., Chen Y., et al. (2023). The eQTL colocalization and transcriptome-wide association study identify potentially causal genes responsible for economic traits in Simmental beef cattle. J. Animal Sci. Biotechnol. 14, 78. 10.1186/s40104-023-00876-7 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

