Transcriptome analysis and immune gene expression of channel catfish (Ictalurus punctatus) fed diets with inclusion of frass from black soldier fly larvae
- PMID: 38264328
- PMCID: PMC10803510
- DOI: 10.3389/fphys.2023.1330368
Transcriptome analysis and immune gene expression of channel catfish (Ictalurus punctatus) fed diets with inclusion of frass from black soldier fly larvae
Abstract
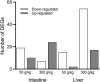
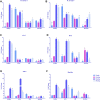
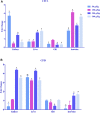
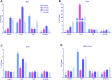
The larval waste, exoskeleton shedding, and leftover feed components of the black soldier fly and its larvae make up the by-product known as frass. In this study, we subjected channel catfish (Ictalurus punctatus) to a 10-week feeding trial to assess how different dietary amounts of frass inclusion would affect both systemic and mucosal tissue gene expression, especially in regard to growth and immune-related genes. Fish were divided in quadruplicate aquaria, and five experimental diets comprising 0, 50, 100, 200, and 300 g of frass per kilogram of feed were fed twice daily. At the end of the trial, liver, head kidney, gill, and intestine samples were collected for gene expression analyses. First, liver and intestine samples from fish fed with a no frass inclusion diet (control), low-frass (50 g/kg) inclusion diet, or a high-frass (300 g/kg) inclusion diet were subjected to Illumina RNA sequencing to determine global differential gene expression among diet groups. Differentially expressed genes (DEGs) included the upregulation of growth-related genes such as glucose-6-phosphatase and myostatin, as well as innate immune receptors and effector molecules such as toll-like receptor 5, apolipoprotein A1, C-type lectin, and lysozyme. Based on the initial screenings of low/high frass using RNA sequencing, a more thorough evaluation of immune gene expression of all tissues sampled, and all levels of frass inclusion, was further conducted. Using targeted quantitative PCR panels for both innate and adaptive immune genes from channel catfish, differential expression of genes was identified, which included innate receptors (TLR1, TLR5, TLR9, and TLR20A), proinflammatory cytokines (IL-1β type a, IL-1β type b, IL-17, IFN-γ, and TNFα), chemokines (CFC3 and CFD), and hepcidin in both systemic (liver and head kidney) and mucosal (gill and intestine) tissues. Overall, frass from black soldier fly larvae inclusion in formulated diets was found to alter global gene expression and activate innate and adaptive immunity in channel catfish, which has the potential to support disease resistance in this species in addition to demonstrated growth benefits.
Keywords: RNA-seq; adaptive immunity; alternative diets; channel catfish; feed additives; frass; innate immunity; metabolism.
Copyright © 2024 Sankappa, Lange, Yildirim-Aksoy, Eljack, Kucuktas, Beck and Abernathy.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Dietary inclusion of frass from black soldier fly (Hermetia illucens) larvae modulates gut microbiota without compromising the growth performance and health status of catfish (Ictalurus punctatus) juveniles.Fish Physiol Biochem. 2025 Aug 5;51(4):137. doi: 10.1007/s10695-025-01552-5. Fish Physiol Biochem. 2025. PMID: 40762807
-
Intestinal histopathology and immune responses following Escherichia coli lipopolysaccharide challenge in Nile tilapia fed enriched black soldier fly larval (BSF) meal supplemented with chitinase.Fish Shellfish Immunol. 2022 Sep;128:620-633. doi: 10.1016/j.fsi.2022.08.050. Epub 2022 Aug 28. Fish Shellfish Immunol. 2022. PMID: 36038101
-
Growth Performance, Survival, Blood Chemistry, and Immune Gene Expression of Channel Catfish (Ictalurus punctatus) Fed Probiotic-Supplemented Diets.Vet Sci. 2022 Dec 16;9(12):701. doi: 10.3390/vetsci9120701. Vet Sci. 2022. PMID: 36548862 Free PMC article.
-
Plasma and tissue transferrin and ferritin, and gene expression of ferritin, transferrin, and transferrin receptors I and II in channel catfish Ictalurus punctatus fed diets with different concentrations of inorganic or organic iron.J Fish Dis. 2024 Aug;47(8):e13953. doi: 10.1111/jfd.13953. Epub 2024 Apr 14. J Fish Dis. 2024. PMID: 38616496
-
Potential Applications of Frass Derived from Black Soldier Fly Larvae Treatment of Food Waste: A Review.Foods. 2022 Sep 1;11(17):2664. doi: 10.3390/foods11172664. Foods. 2022. PMID: 36076850 Free PMC article. Review.
Cited by
-
Impact of mediterranean fruit fly rearing residues and biological supplementation on performance of gimmizah chicks.Poult Sci. 2025 Jul;104(7):105198. doi: 10.1016/j.psj.2025.105198. Epub 2025 Apr 19. Poult Sci. 2025. PMID: 40294550 Free PMC article.
-
Black Soldier Fly Larvae as a Novel Protein Feed Resource Promoting Circular Economy in Agriculture.Insects. 2025 Aug 10;16(8):830. doi: 10.3390/insects16080830. Insects. 2025. PMID: 40870631 Free PMC article. Review.
References
-
- Adeoye A. A., Akegbejo‐Samsons Y., Fawole F. J., Davies S. J. (2020). Preliminary assessment of black soldier fly (Hermetia illucens) larval meal in the diet of African catfish (Clarias gariepinus): impact on growth, body index, and hematological parameters. J. World Aquac. Soc. 51, 1024–1033. 10.1111/jwas.12691 - DOI
-
- Agbohessou P. S., Mandiki S. N. M., Biyong S. R. M., Cornet V., Nguyen T. M., Lambert J., et al. (2022). Intestinal histopathology and immune responses following Escherichia coli lipopolysaccharide challenge in Nile tilapia fed enriched black soldier fly larval (BSF) meal supplemented with chitinase. Fish. Shellfish Immunol. 128, 620–633. 10.1016/j.fsi.2022.08.050 - DOI - PubMed
-
- Andrews S. (2017). FastQC: a quality control tool for high throughput sequence data. 2010.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

