LATTE: Label-efficient incident phenotyping from longitudinal electronic health records
- PMID: 38264714
- PMCID: PMC10801250
- DOI: 10.1016/j.patter.2023.100906
LATTE: Label-efficient incident phenotyping from longitudinal electronic health records
Abstract
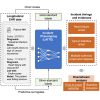
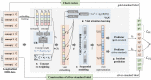
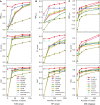
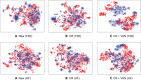
Electronic health record (EHR) data are increasingly used to support real-world evidence studies but are limited by the lack of precise timings of clinical events. Here, we propose a label-efficient incident phenotyping (LATTE) algorithm to accurately annotate the timing of clinical events from longitudinal EHR data. By leveraging the pre-trained semantic embeddings, LATTE selects predictive features and compresses their information into longitudinal visit embeddings through visit attention learning. LATTE models the sequential dependency between the target event and visit embeddings to derive the timings. To improve label efficiency, LATTE constructs longitudinal silver-standard labels from unlabeled patients to perform semi-supervised training. LATTE is evaluated on the onset of type 2 diabetes, heart failure, and relapses of multiple sclerosis. LATTE consistently achieves substantial improvements over benchmark methods while providing high prediction interpretability. The event timings are shown to help discover risk factors of heart failure among patients with rheumatoid arthritis.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures

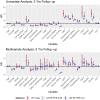
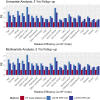
References
-
- Ananthakrishnan A.N., Cai T., Savova G., Cheng S.C., Chen P., Perez R.G., Gainer V.S., Murphy S.N., Szolovits P., Xia Z., et al. Improving case definition of crohn’s disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach. Inflamm. Bowel Dis. 2013;19:1411–1420. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

