Etiology of lower respiratory tract in pneumonia based on metagenomic next-generation sequencing: a retrospective study
- PMID: 38264726
- PMCID: PMC10803656
- DOI: 10.3389/fcimb.2023.1291980
Etiology of lower respiratory tract in pneumonia based on metagenomic next-generation sequencing: a retrospective study
Abstract
Introduction: Pneumonia are the leading cause of death worldwide, and antibiotic treatment remains fundamental. However, conventional sputum smears or cultures are still inefficient for obtaining pathogenic microorganisms.Metagenomic next-generation sequencing (mNGS) has shown great value in nucleic acid detection, however, the NGS results for lower respiratory tract microorganisms are still poorly studied.
Methods: This study dealt with investigating the efficacy of mNGS in detecting pathogens in the lower respiratory tract of patients with pulmonary infections. A total of 112 patients admitted at the First Affiliated Hospital of Zhengzhou University between April 30, 2018, and June 30, 2020, were enrolled in this retrospective study. The bronchoalveolar lavage fluid (BALF) was obtained from lower respiratory tract from each patient. Routine methods (bacterial smear and culture) and mNGS were employed for the identification of pathogenic microorganisms in BALF.
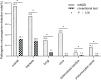
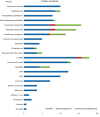
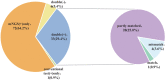
Results: The average patient age was 53.0 years, with 94.6% (106/112) obtaining pathogenic microorganism results. The total mNGS detection rate of pathogenic microorganisms significantly surpassed conventional methods (93.7% vs. 32.1%, P < 0.05). Notably, 75% of patients (84/112) were found to have bacteria by mNGS, but only 28.6% (32/112) were found to have bacteria by conventional approaches. The most commonly detected bacteria included Acinetobacter baumannii (19.6%), Klebsiella pneumoniae (17.9%), Pseudomonas aeruginosa (14.3%), Staphylococcus faecium (12.5%), Enterococcus faecium (12.5%), and Haemophilus parainfluenzae (11.6%). In 29.5% (33/112) of patients, fungi were identified using mNGS, including 23 cases of Candida albicans (20.5%), 18 of Pneumocystis carinii (16.1%), and 10 of Aspergillus (8.9%). However, only 7.1 % (8/112) of individuals were found to have fungi when conventional procedures were used. The mNGS detection rate of viruses was significantly higher than the conventional method rate (43.8% vs. 0.9%, P < 0.05). The most commonly detected viruses included Epstein-Barr virus (15.2%), cytomegalovirus (13.4%), circovirus (8.9%), human coronavirus (4.5%), and rhinovirus (4.5%). Only 29.4% (33/112) of patients were positive, whereas 5.4% (6/112) of patients were negative for both detection methods as shown by Kappa analysis, indicating poor consistency between the two methods (P = 0.340; Kappa analysis).
Conclusion: Significant benefits of mNGS have been shown in the detection of pathogenic microorganisms in patients with pulmonary infection. For those with suboptimal therapeutic responses, mNGS can provide an etiological basis, aiding in precise anti-infective treatment.
Keywords: antibiotics; etiology; lower respiratory tract; metagenomic next-generation sequencing; pulmonary infection.
Copyright © 2024 Wang, Yuan, Yang, Sun, Hou, Zhang and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
[Microbiological characteristics of patients with severe burns caused by blast and application of meta- genomics next-generation sequencing in the detection of pathogenic microorganisms].Zhonghua Shao Shang Za Zhi. 2021 Oct 20;37(10):946-952. doi: 10.3760/cma.j.cn501120-20201017-00440. Zhonghua Shao Shang Za Zhi. 2021. PMID: 34689464 Free PMC article. Chinese.
-
[Diagnostic value of detection of pathogens in bronchoalveolar lavage fluid by metagenomics next-generation sequencing in organ transplant patients with pulmonary infection].Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2021 Dec;33(12):1440-1446. doi: 10.3760/cma.j.cn121430-20211008-01439. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2021. PMID: 35131010 Chinese.
-
Metagenomic next-generation sequencing of bronchoalveolar lavage fluid from children with severe pneumonia in pediatric intensive care unit.Front Cell Infect Microbiol. 2023 Mar 16;13:1082925. doi: 10.3389/fcimb.2023.1082925. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37009495 Free PMC article.
-
Metagenomic next generation sequencing of bronchoalveolar lavage fluids for the identification of pathogens in patients with pulmonary infection: A retrospective study.Diagn Microbiol Infect Dis. 2024 Sep;110(1):116402. doi: 10.1016/j.diagmicrobio.2024.116402. Epub 2024 Jun 12. Diagn Microbiol Infect Dis. 2024. PMID: 38878340 Review.
-
Metagenomic next-generation sequencing on treatment strategies and prognosis of patients with lower respiratory tract infections: A systematic review and meta-analysis.Int J Antimicrob Agents. 2025 Mar;65(3):107440. doi: 10.1016/j.ijantimicag.2024.107440. Epub 2025 Jan 4. Int J Antimicrob Agents. 2025. PMID: 39761759
Cited by
-
Consistency between metagenomic next-generation sequencing versus traditional microbiological tests for infective disease: systemic review and meta-analysis.Crit Care. 2025 Feb 3;29(1):55. doi: 10.1186/s13054-025-05288-9. Crit Care. 2025. PMID: 39901264 Free PMC article.
-
Optimization of 18 S rRNA metabarcoding for the simultaneous diagnosis of intestinal parasites.Sci Rep. 2024 Oct 23;14(1):25049. doi: 10.1038/s41598-024-76304-1. Sci Rep. 2024. PMID: 39443558 Free PMC article.
-
The clinical application of metagenomic next-generation sequencing in immunocompromised patients with severe respiratory infections in the ICU.Respir Res. 2024 Oct 5;25(1):360. doi: 10.1186/s12931-024-02991-z. Respir Res. 2024. PMID: 39369191 Free PMC article.
-
Antimicrobial strategies of lower respiratory tract infections in immunocompromised patients based on metagenomic next-generation sequencing: a retrospective study.BMC Infect Dis. 2025 Mar 14;25(1):360. doi: 10.1186/s12879-025-10753-5. BMC Infect Dis. 2025. PMID: 40087607 Free PMC article.
-
Comparison of the Impact of tNGS with mNGS on Antimicrobial Management in Patients with LRTIs: A Multicenter Retrospective Cohort Study.Infect Drug Resist. 2025 Jan 6;18:93-105. doi: 10.2147/IDR.S493575. eCollection 2025. Infect Drug Resist. 2025. PMID: 39803312 Free PMC article.
References
-
- Boldrin E., Mazza M., Piano M. A., Alfieri R., Montagner I. M., Magni G., et al. . (2022). Putative Clinical Potential of ERBB2 Amplification Assessment by ddPCR in FFPE-DNA and cfDNA of Gastroesophageal Adenocarcinoma Patients. Cancers (Basel). 14 (9), 2180. doi: 10.3390/cancers14092180 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

