Universal dynamic fitting of magnetic resonance spectroscopy
- PMID: 38265152
- PMCID: PMC7616727
- DOI: 10.1002/mrm.30001
Universal dynamic fitting of magnetic resonance spectroscopy
Abstract
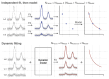
Purpose: Dynamic (2D) MRS is a collection of techniques where acquisitions of spectra are repeated under varying experimental or physiological conditions. Dynamic MRS comprises a rich set of contrasts, including diffusion-weighted, relaxation-weighted, functional, edited, or hyperpolarized spectroscopy, leading to quantitative insights into multiple physiological or microstructural processes. Conventional approaches to dynamic MRS analysis ignore the shared information between spectra, and instead proceed by independently fitting noisy individual spectra before modeling temporal changes in the parameters. Here, we propose a universal dynamic MRS toolbox which allows simultaneous fitting of dynamic spectra of arbitrary type.
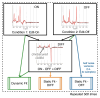
Methods: A simple user-interface allows information to be shared and precisely modeled across spectra to make inferences on both spectral and dynamic processes. We demonstrate and thoroughly evaluate our approach in three types of dynamic MRS techniques. Simulations of functional and edited MRS are used to demonstrate the advantages of dynamic fitting.
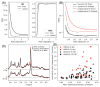
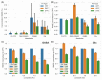
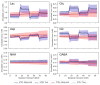
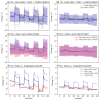
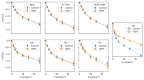
Results: Analysis of synthetic functional 1H-MRS data shows a marked decrease in parameter uncertainty as predicted by prior work. Analysis with our tool replicates the results of two previously published studies using the original in vivo functional and diffusion-weighted data. Finally, joint spectral fitting with diffusion orientation models is demonstrated in synthetic data.
Conclusion: A toolbox for generalized and universal fitting of dynamic, interrelated MR spectra has been released and validated. The toolbox is shared as a fully open-source software with comprehensive documentation, example data, and tutorials.
Keywords: MRS; dMRS; edited‐MRS; fMRS; spectroscopy.
© 2024 The Authors. Magnetic Resonance in Medicine published by Wiley Periodicals LLC on behalf of International Society for Magnetic Resonance in Medicine.
Figures



References
-
- Craven AR, Dwyer G, Ersland L, et al. GABA, glutamatergic dynamics and BOLD contrast concurrently assessed using functional MR spectroscopy during a cognitive task. bioRxiv. 2023;2023:539017. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

