Measures of genetic diversification in somatic tissues at bulk and single-cell resolution
- PMID: 38265286
- PMCID: PMC10945735
- DOI: 10.7554/eLife.89780
Measures of genetic diversification in somatic tissues at bulk and single-cell resolution
Abstract
Intra-tissue genetic heterogeneity is universal to both healthy and cancerous tissues. It emerges from the stochastic accumulation of somatic mutations throughout development and homeostasis. By combining population genetics theory and genomic information, genetic heterogeneity can be exploited to infer tissue organization and dynamics in vivo. However, many basic quantities, for example the dynamics of tissue-specific stem cells remain difficult to quantify precisely. Here, we show that single-cell and bulk sequencing data inform on different aspects of the underlying stochastic processes. Bulk-derived variant allele frequency spectra (VAF) show transitions from growing to constant stem cell populations with age in samples of healthy esophagus epithelium. Single-cell mutational burden distributions allow a sample size independent measure of mutation and proliferation rates. Mutation rates in adult hematopietic stem cells are higher compared to inferences during development, suggesting additional proliferation-independent effects. Furthermore, single-cell derived VAF spectra contain information on the number of tissue-specific stem cells. In hematopiesis, we find approximately 2 × 105 HSCs, if all stem cells divide symmetrically. However, the single-cell mutational burden distribution is over-dispersed compared to a model of Poisson distributed random mutations. A time-associated model of mutation accumulation with a constant rate alone cannot generate such a pattern. At least one additional source of stochasticity would be needed. Possible candidates for these processes may be occasional bursts of stem cell divisions, potentially in response to injury, or non-constant mutation rates either through environmental exposures or cell-intrinsic variation.
Keywords: evolutionary biology; evolutionary inferences; healthy human tissues; human; sampling; single-cell mutation burden; stem cell dynamics; varient allele frequency.
© 2023, Moeller, Mon Père et al.
Conflict of interest statement
MM, NM, BW, WH No competing interests declared
Figures
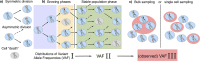





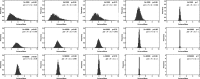


Update of
- doi: 10.1101/2022.11.07.515470
- doi: 10.7554/eLife.89780.1
- doi: 10.7554/eLife.89780.2
Comment in
- doi: 10.7554/eLife.95513
References
-
- Abascal F, Harvey LMR, Mitchell E, Lawson ARJ, Lensing SV, Ellis P, Russell AJC, Alcantara RE, Baez-Ortega A, Wang Y, Kwa EJ, Lee-Six H, Cagan A, Coorens THH, Chapman MS, Olafsson S, Leonard S, Jones D, Machado HE, Davies M, Øbro NF, Mahubani KT, Allinson K, Gerstung M, Saeb-Parsy K, Kent DG, Laurenti E, Stratton MR, Rahbari R, Campbell PJ, Osborne RJ, Martincorena I. Somatic mutation landscapes at single-molecule resolution. Nature. 2021;593:405–410. doi: 10.1038/s41586-021-03477-4. - DOI - PubMed
-
- Bailey C, Black JRM, Reading JL, Litchfield K, Turajlic S, McGranahan N, Jamal-Hanjani M, Swanton C. Tracking cancer evolution through the disease course. Cancer Discovery. 2021;11:916–932. doi: 10.1158/2159-8290.CD-20-1559. - DOI - PMC - PubMed
-
- Cagan A, Baez-Ortega A, Brzozowska N, Abascal F, Coorens THH, Sanders MA, Lawson ARJ, Harvey LMR, Bhosle S, Jones D, Alcantara RE, Butler TM, Hooks Y, Roberts K, Anderson E, Lunn S, Flach E, Spiro S, Januszczak I, Wrigglesworth E, Jenkins H, Dallas T, Masters N, Perkins MW, Deaville R, Druce M, Bogeska R, Milsom MD, Neumann B, Gorman F, Constantino-Casas F, Peachey L, Bochynska D, Smith ESJ, Gerstung M, Campbell PJ, Murchison EP, Stratton MR, Martincorena I. Somatic mutation rates scale with lifespan across mammals. Nature. 2022;604:517–524. doi: 10.1038/s41586-022-04618-z. - DOI - PMC - PubMed
-
- Caravagna G, Heide T, Williams MJ, Zapata L, Nichol D, Chkhaidze K, Cross W, Cresswell GD, Werner B, Acar A, Chesler L, Barnes CP, Sanguinetti G, Graham TA, Sottoriva A. Subclonal reconstruction of tumors by using machine learning and population genetics. Nature Genetics. 2020;52:898–907. doi: 10.1038/s41588-020-0675-5. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

