Meta-analysis reveals obesity associated gut microbial alteration patterns and reproducible contributors of functional shift
- PMID: 38265338
- PMCID: PMC10810176
- DOI: 10.1080/19490976.2024.2304900
Meta-analysis reveals obesity associated gut microbial alteration patterns and reproducible contributors of functional shift
Abstract
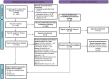
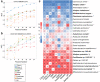
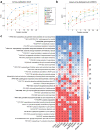
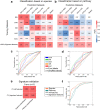
The majority of cohort-specific studies associating gut microbiota with obesity are often contradictory; thus, the replicability of the signature remains questionable. Moreover, the species that drive obesity-associated functional shifts and their replicability remain unexplored. Thus, we aimed to address these questions by analyzing gut microbial metagenome sequencing data to develop an in-depth understanding of obese host-gut microbiota interactions using 3329 samples (Obese, n = 1494; Control, n = 1835) from 17 different countries, including both 16S rRNA gene and metagenomic sequence data. Fecal metagenomic data from diverse geographical locations were curated, profiled, and pooled using a machine learning-based approach to identify robust global signatures of obesity. Furthermore, gut microbial species and pathways were systematically integrated through the genomic content of the species to identify contributors to obesity-associated functional shifts. The community structure of the obese gut microbiome was evaluated, and a reproducible depletion of diversity was observed in the obese compared to the lean gut. From this, we infer that the loss of diversity in the obese gut is responsible for perturbations in the healthy microbial functional repertoire. We identified 25 highly predictive species and 37 pathway associations as signatures of obesity, which were validated with remarkably high accuracy (AUC, Species: 0.85, and pathway: 0.80) with an independent validation dataset. We observed a reduction in short-chain fatty acid (SCFA) producers (several Alistipes species, Odoribacter splanchnicus, etc.) and depletion of promoters of gut barrier integrity (Akkermansia muciniphila and Bifidobacterium longum) in obese guts. Our analysis underlines SCFAs and purine/pyrimidine biosynthesis, carbohydrate metabolism pathways in control individuals, and amino acid, enzyme cofactor, and peptidoglycan biosynthesis pathway enrichment in obese individuals. We also mapped the contributors to important obesity-associated functional shifts and observed that these are both dataset-specific and shared across the datasets. In summary, a comprehensive analysis of diverse datasets unveils species specifically contributing to functional shifts and consistent gut microbial patterns associated to obesity.
Keywords: functional contributors; gut microbial association; machine learning; meta-analysis; obesity.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- WHO . 2021. Obesity and overweight. http://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight.
-
- Wheeler E, Huang N, Bochukova EG, Keogh JM, Lindsay S, Garg S, Henning E, Blackburn H, Loos RJ, Wareham NJ. et al. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat Genet. 2013;45(5):513–517. doi:10.1038/NG.2607. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
