Phenotypic and genotypic characterization of Marinobacterium weihaiense sp. nov. and Marinobacterium marinum sp. nov., isolated from marine sediment, and genomic properties of the genus Marinobacterium
- PMID: 38265428
- PMCID: PMC10868613
- DOI: 10.1099/mgen.0.001182
Phenotypic and genotypic characterization of Marinobacterium weihaiense sp. nov. and Marinobacterium marinum sp. nov., isolated from marine sediment, and genomic properties of the genus Marinobacterium
Abstract
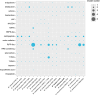
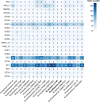
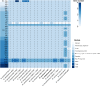
In this study, two novel bacterial strains were isolated from coastal sediment of Weihai, China. The two strains were Gram-stain-negative and facultatively aerobic, designated 3-1745T and A346T. Based on phenotypic, genetic and phylogenetic properties, strains 3-1745T and A346T represent two novel species of the genus Marinobacterium. The results of genome analysis revealed many central carbohydrate metabolism pathways such as gluconeogenesis, pyruvate oxidation, tricyclic acid cycle, pentose phosphate pathway and PRPP biosynthesis in the genus Marinobacterium. The ability of strains 3-1745T and A346T to utilize volatile fatty acids was experimentally confirmed. Polyhydroxyalkanoate synthases (PhaA, PhaB and PhaC) for the synthesis of polyhydroxyalkanoates were prevalent in the genus Marinobacterium. Multiple BGCs (biosynthetic gene clusters) including betalactone, ectoine, ranthipeptide, redox-cofactor, RiPPs (ribosomally synthesized post-translationally modified peptides) and T3PKS (polyketide synthases) in the genome of the genus Marinobacterium were found. Additional genome analyses suggested that the genus Marinobacterium contained diverse potential mechanisms of salt tolerance and mainly utilized oligosaccharides. This is the first report on broad genomic analyses of the genus Marinobacterium with the description of two novel species and potential ecological and biotechnological implications.
Keywords: Marinobacterium; comparative genomic analysis; polyhydroxyalkanoates; polyphasic taxonomy.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Marinobacterium nitratireducens sp. nov. and Marinobacterium sediminicola sp. nov., isolated from marine sediment.Int J Syst Evol Microbiol. 2009 May;59(Pt 5):1173-8. doi: 10.1099/ijs.0.005751-0. Int J Syst Evol Microbiol. 2009. PMID: 19406814
-
Marinobacterium aestuarii sp. nov., a benzene-degrading marine bacterium isolated from estuary sediment.Int J Syst Evol Microbiol. 2018 Feb;68(2):651-656. doi: 10.1099/ijsem.0.002561. Epub 2018 Jan 5. Int J Syst Evol Microbiol. 2018. PMID: 29303694
-
Genomic insights into Marinobacterium sediminicola CGMCC 1.7287T: A polyhydroxyalkanoate-producing bacterium isolated from marine sediment.Mar Genomics. 2025 Apr 15;80:101180. doi: 10.1016/j.margen.2025.101180. Epub 2025 Feb 12. Mar Genomics. 2025. PMID: 39993876
-
Marinobacterium rhizophilum sp. nov., isolated from the rhizosphere of the coastal tidal-flat plant Suaeda japonica.Int J Syst Evol Microbiol. 2008 Jan;58(Pt 1):164-7. doi: 10.1099/ijs.0.65176-0. Int J Syst Evol Microbiol. 2008. PMID: 18175703
-
Marinobacterium marisflavi sp. nov., isolated from a costal seawater.Curr Microbiol. 2009 May;58(5):511-5. doi: 10.1007/s00284-009-9355-5. Epub 2009 Feb 3. Curr Microbiol. 2009. PMID: 19189183
Cited by
-
Validation List no. 221: valid publication of new names and new combinations effectively published outside the IJSEM.Int J Syst Evol Microbiol. 2025 Jan;75(1):006562. doi: 10.1099/ijsem.0.006562. Int J Syst Evol Microbiol. 2025. PMID: 39887304 Free PMC article. No abstract available.
References
-
- González JM, Mayer F, Moran MA, Hodson RE, Whitman WB. Microbulbifer hydrolyticus gen. nov., sp. nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int J Syst Bacteriol. 1997;47:369–376. doi: 10.1099/00207713-47-2-369. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

