Mycobacterium tuberculosis response to cholesterol is integrated with environmental pH and potassium levels via a lipid metabolism regulator
- PMID: 38266039
- PMCID: PMC10843139
- DOI: 10.1371/journal.pgen.1011143
Mycobacterium tuberculosis response to cholesterol is integrated with environmental pH and potassium levels via a lipid metabolism regulator
Abstract
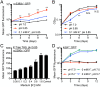
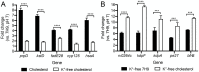
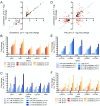
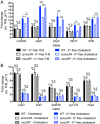
Successful colonization of the host requires Mycobacterium tuberculosis (Mtb) to sense and respond coordinately to disparate environmental cues during infection and adapt its physiology. However, how Mtb response to environmental cues and the availability of key carbon sources may be integrated is poorly understood. Here, by exploiting a reporter-based genetic screen, we have unexpectedly found that overexpression of transcription factors involved in Mtb lipid metabolism altered the dampening effect of low environmental potassium concentrations ([K+]) on the pH response of Mtb. Cholesterol is a major carbon source for Mtb during infection, and transcriptional analyses revealed that Mtb response to acidic pH was augmented in the presence of cholesterol and vice versa. Strikingly, deletion of the putative lipid regulator mce3R had little effect on Mtb transcriptional response to acidic pH or cholesterol individually, but resulted specifically in loss of cholesterol response augmentation in the simultaneous presence of acidic pH. Similarly, while mce3R deletion had little effect on Mtb response to low environmental [K+] alone, augmentation of the low [K+] response by the simultaneous presence of cholesterol was lost in the mutant. Finally, a mce3R deletion mutant was attenuated for growth in foamy macrophages and for colonization in a murine infection model that recapitulates caseous necrotic lesions and the presence of foamy macrophages. These findings reveal the critical coordination between Mtb response to environmental cues and cholesterol, a vital carbon source, and establishes Mce3R as a transcription factor that crucially serves to integrate these signals.
Copyright: © 2024 Chen et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Update of
-
Mycobacterium tuberculosis response to cholesterol is integrated with environmental pH and potassium levels via a lipid utilization regulator.bioRxiv [Preprint]. 2023 Aug 24:2023.08.22.554309. doi: 10.1101/2023.08.22.554309. bioRxiv. 2023. Update in: PLoS Genet. 2024 Jan 24;20(1):e1011143. doi: 10.1371/journal.pgen.1011143. PMID: 37662244 Free PMC article. Updated. Preprint.
Similar articles
-
Mycobacterium tuberculosis response to cholesterol is integrated with environmental pH and potassium levels via a lipid utilization regulator.bioRxiv [Preprint]. 2023 Aug 24:2023.08.22.554309. doi: 10.1101/2023.08.22.554309. bioRxiv. 2023. Update in: PLoS Genet. 2024 Jan 24;20(1):e1011143. doi: 10.1371/journal.pgen.1011143. PMID: 37662244 Free PMC article. Updated. Preprint.
-
Cholesterol metabolism and intrabacterial potassium homeostasis are intrinsically related in Mycobacterium tuberculosis.PLoS Pathog. 2025 May 22;21(5):e1013207. doi: 10.1371/journal.ppat.1013207. eCollection 2025 May. PLoS Pathog. 2025. PMID: 40402977 Free PMC article.
-
Slow growth of Mycobacterium tuberculosis at acidic pH is regulated by phoPR and host-associated carbon sources.Mol Microbiol. 2014 Oct;94(1):56-69. doi: 10.1111/mmi.12688. Epub 2014 Jul 13. Mol Microbiol. 2014. PMID: 24975990 Free PMC article.
-
Targeting Mycobacterium tuberculosis pH-driven adaptation.Microbiology (Reading). 2024 May;170(5):001458. doi: 10.1099/mic.0.001458. Microbiology (Reading). 2024. PMID: 38717801 Free PMC article. Review.
-
Role of cholesterol in Mycobacterium tuberculosis infection.Indian J Exp Biol. 2009 Jun;47(6):407-11. Indian J Exp Biol. 2009. PMID: 19634704 Review.
Cited by
-
Unraveling Mycobacterium tuberculosis acid resistance and pH homeostasis mechanisms.FEBS Lett. 2025 Jun;599(12):1634-1648. doi: 10.1002/1873-3468.70023. Epub 2025 Feb 24. FEBS Lett. 2025. PMID: 39995204 Free PMC article. Review.
-
Mycobacterium tuberculosis Mce3R TetR-like Repressor Forms an Asymmetric Four-Helix Bundle and Binds a Nonpalindrome Sequence†.ACS Chem Biol. 2024 Dec 20;19(12):2580-2592. doi: 10.1021/acschembio.4c00687. Epub 2024 Nov 15. ACS Chem Biol. 2024. PMID: 39545866 Free PMC article.
-
Building Spatiotemporal Understanding of Mycobacterium tuberculosis-Host Interactions.ACS Infect Dis. 2025 Feb 14;11(2):277-286. doi: 10.1021/acsinfecdis.4c00840. Epub 2025 Jan 23. ACS Infect Dis. 2025. PMID: 39847659 Review.
-
Uncovering the roles of Mycobacterium tuberculosis melH in redox and bioenergetic homeostasis: implications for antitubercular therapy.mSphere. 2024 Apr 23;9(4):e0006124. doi: 10.1128/msphere.00061-24. Epub 2024 Apr 2. mSphere. 2024. PMID: 38564709 Free PMC article.
-
Cholesterol metabolism and intrabacterial potassium homeostasis are intrinsically related in Mycobacterium tuberculosis.bioRxiv [Preprint]. 2024 Nov 11:2024.11.10.622811. doi: 10.1101/2024.11.10.622811. bioRxiv. 2024. Update in: PLoS Pathog. 2025 May 22;21(5):e1013207. doi: 10.1371/journal.ppat.1013207. PMID: 39605342 Free PMC article. Updated. Preprint.
References
-
- World Health Organization. Global tuberculosis report 2023. https://www.who.int/publications/i/item/9789240083851 (last accessed 11 January 2024).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

