Sparse species interactions reproduce abundance correlation patterns in microbial communities
- PMID: 38266051
- PMCID: PMC10853627
- DOI: 10.1073/pnas.2309575121
Sparse species interactions reproduce abundance correlation patterns in microbial communities
Abstract
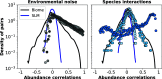
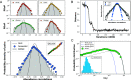
During the last decades, macroecology has identified broad-scale patterns of abundances and diversity of microbial communities and put forward some potential explanations for them. However, these advances are not paralleled by a full understanding of the dynamical processes behind them. In particular, abundance fluctuations of different species are found to be correlated, both across time and across communities in metagenomic samples. Reproducing such correlations through appropriate population models remains an open challenge. The present paper tackles this problem and points to sparse species interactions as a necessary mechanism to account for them. Specifically, we discuss several possibilities to include interactions in population models and recognize Lotka-Volterra constants as a successful ansatz. For this, we design a Bayesian inference algorithm to extract sets of interaction constants able to reproduce empirical probability distributions of pairwise correlations for diverse biomes. Importantly, the inferred models still reproduce well-known single-species macroecological patterns concerning abundance fluctuations across both species and communities. Endorsed by the agreement with the empirically observed phenomenology, our analyses provide insights into the properties of the networks of microbial interactions, revealing that sparsity is a crucial feature.
Keywords: Lotka–Volterra; correlations; ecology; interactions; microbiome.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


Similar articles
-
Environmental fluctuations explain the universal decay of species-abundance correlations with phylogenetic distance.Proc Natl Acad Sci U S A. 2023 Sep 12;120(37):e2217144120. doi: 10.1073/pnas.2217144120. Epub 2023 Sep 5. Proc Natl Acad Sci U S A. 2023. PMID: 37669363 Free PMC article.
-
MetaMIS: a metagenomic microbial interaction simulator based on microbial community profiles.BMC Bioinformatics. 2016 Nov 25;17(1):488. doi: 10.1186/s12859-016-1359-0. BMC Bioinformatics. 2016. PMID: 27887570 Free PMC article.
-
Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression.PLoS One. 2014 Jul 23;9(7):e102451. doi: 10.1371/journal.pone.0102451. eCollection 2014. PLoS One. 2014. PMID: 25054627 Free PMC article.
-
Practical considerations for sampling and data analysis in contemporary metagenomics-based environmental studies.J Microbiol Methods. 2018 Nov;154:14-18. doi: 10.1016/j.mimet.2018.09.020. Epub 2018 Oct 1. J Microbiol Methods. 2018. PMID: 30287354 Review.
-
Metagenome-scale community metabolic modelling for understanding the role of gut microbiota in human health.Comput Biol Med. 2022 Oct;149:105997. doi: 10.1016/j.compbiomed.2022.105997. Epub 2022 Aug 19. Comput Biol Med. 2022. PMID: 36055158 Review.
Cited by
-
Macroecological patterns in experimental microbial communities.PLoS Comput Biol. 2025 May 8;21(5):e1013044. doi: 10.1371/journal.pcbi.1013044. eCollection 2025 May. PLoS Comput Biol. 2025. PMID: 40341906 Free PMC article.
-
A macroecological perspective on genetic diversity in the human gut microbiome.PLoS One. 2023 Jul 21;18(7):e0288926. doi: 10.1371/journal.pone.0288926. eCollection 2023. PLoS One. 2023. PMID: 37478102 Free PMC article.
-
Exploring interspecific interaction variability in microbiota: A review.Eng Microbiol. 2024 Nov 9;4(4):100178. doi: 10.1016/j.engmic.2024.100178. eCollection 2024 Dec. Eng Microbiol. 2024. PMID: 40104221 Free PMC article. Review.
References
-
- Handelsman J., Rondon M. R., Brady S. F., Clardy J., Goodman R. M., Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 5, R245–R249 (1998). - PubMed
-
- Steele H. L., Streit W. R., Metagenomics: Advances in ecology and biotechnology. FEMS Microbiol. Lett. 247, 105–111 (2005). - PubMed
-
- Rappé M. S., Giovannoni S. J., The uncultured microbial majority. Annu. Rev. Microbiol. 57, 369–394 (2003). - PubMed
-
- Hug L. A., et al. , A new view of the tree of life. Nat. Microbiol. 1, 16048 (2016). - PubMed
MeSH terms
Grants and funding
- PGC2018-098186-B-I00/Ministerio de Ciencia e Innovación (MCIN)
- PID2021-128966NB-I00/Ministerio de Ciencia e Innovación (MCIN)
- PID2020-113681GB-I00/Ministerio de Ciencia e Innovación (MCIN)
- B-FQM-366-UGR20/Consejería de Conocimiento, Investigación y Universidad, Junta de Andalucía (Ministry of Knowledge, Research and University, Andalusia)
- PGC2022-141802NB-I00/Ministerio de Ciencia e Innovación (MCIN)
LinkOut - more resources
Full Text Sources

