DNA Methylation-Based Assessment of Cell Composition in Human Pancreas and Islets
- PMID: 38266068
- PMCID: PMC10958580
- DOI: 10.2337/db23-0704
DNA Methylation-Based Assessment of Cell Composition in Human Pancreas and Islets
Abstract
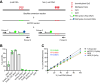
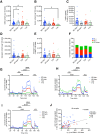
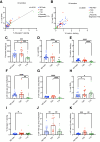
Assessment of pancreas cell type composition is crucial to the understanding of the genesis of diabetes. Current approaches use immunodetection of protein markers, for example, insulin as a marker of β-cells. A major limitation of these methods is that protein content varies in physiological and pathological conditions, complicating the extrapolation to actual cell number. Here, we demonstrate the use of cell type-specific DNA methylation markers for determining the fraction of specific cell types in human islet and pancreas specimens. We identified genomic loci that are uniquely demethylated in specific pancreatic cell types and applied targeted PCR to assess the methylation status of these loci in tissue samples, enabling inference of cell type composition. In islet preparations, normalization of insulin secretion to β-cell DNA revealed similar β-cell function in pre-type 1 diabetes (T1D), T1D, and type 2 diabetes (T2D), which was significantly lower than in donors without diabetes. In histological pancreas specimens from recent-onset T1D, this assay showed β-cell fraction within the normal range, suggesting a significant contribution of β-cell dysfunction. In T2D pancreata, we observed increased α-cell fraction and normal β-cell fraction. Methylation-based analysis provides an accurate molecular alternative to immune detection of cell types in the human pancreas, with utility in the interpretation of insulin secretion assays and the assessment of pancreas cell composition in health and disease.
© 2024 by the American Diabetes Association.
Conflict of interest statement
Figures


References
-
- Leete P, Willcox A, Krogvold L, et al. . Differential insulitic profiles determine the extent of β-cell destruction and the age at onset of type 1 diabetes. Diabetes 2016;65:1362–1369 - PubMed
-
- Aly H, Gottlieb P.. The honeymoon phase: intersection of metabolism and immunology. Curr Opin Endocrinol Diabetes Obes 2009;16:286–292 - PubMed
-
- Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC.. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes 2003;52:102–110 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

