Beyond antibiotic resistance: The whiB7 transcription factor coordinates an adaptive response to alanine starvation in mycobacteria
- PMID: 38266648
- PMCID: PMC11031301
- DOI: 10.1016/j.chembiol.2023.12.020
Beyond antibiotic resistance: The whiB7 transcription factor coordinates an adaptive response to alanine starvation in mycobacteria
Abstract
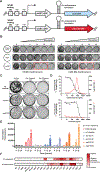
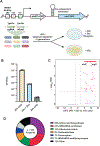
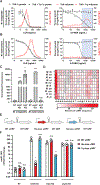
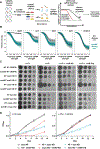
Pathogenic mycobacteria are a significant cause of morbidity and mortality worldwide. The conserved whiB7 stress response reduces the effectiveness of antibiotic therapy by activating several intrinsic antibiotic resistance mechanisms. Despite our comprehensive biochemical understanding of WhiB7, the complex set of signals that induce whiB7 expression remain less clear. We employed a reporter-based, genome-wide CRISPRi epistasis screen to identify a diverse set of 150 mycobacterial genes whose inhibition results in constitutive whiB7 expression. We show that whiB7 expression is determined by the amino acid composition of the 5' regulatory uORF, thereby allowing whiB7 to sense amino acid starvation. Although deprivation of many amino acids can induce whiB7, whiB7 specifically coordinates an adaptive response to alanine starvation by engaging in a feedback loop with the alanine biosynthetic enzyme, aspC. These findings describe a metabolic function for whiB7 and help explain its evolutionary conservation across mycobacterial species occupying diverse ecological niches.
Keywords: CRISPRi; WhiB7; amino acids; antibiotics; mycobacteria; stress response; translation; tuberculosis.
Copyright © 2023 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
Update of
-
Beyond antibiotic resistance: the whiB7 transcription factor coordinates an adaptive response to alanine starvation in mycobacteria.bioRxiv [Preprint]. 2023 Jun 5:2023.06.02.543512. doi: 10.1101/2023.06.02.543512. bioRxiv. 2023. Update in: Cell Chem Biol. 2024 Apr 18;31(4):669-682.e7. doi: 10.1016/j.chembiol.2023.12.020. PMID: 37333137 Free PMC article. Updated. Preprint.
References
-
- World Health Organisation (2021). WHO Global Tuberculosis Report 2021.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

