Interleukin 21 Drives a Hypermetabolic State and CD4+ T-Cell-Associated Pathogenicity in Chronic Intestinal Inflammation
- PMID: 38266738
- PMCID: PMC11034723
- DOI: 10.1053/j.gastro.2024.01.026
Interleukin 21 Drives a Hypermetabolic State and CD4+ T-Cell-Associated Pathogenicity in Chronic Intestinal Inflammation
Abstract
Background & aims: Incapacitated regulatory T cells (Tregs) contribute to immune-mediated diseases. Inflammatory Tregs are evident during human inflammatory bowel disease; however, mechanisms driving the development of these cells and their function are not well understood. Therefore, we investigated the role of cellular metabolism in Tregs relevant to gut homeostasis.
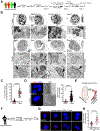
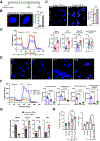
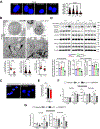
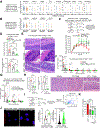
Methods: Using human Tregs, we performed mitochondrial ultrastructural studies via electron microscopy and confocal imaging, biochemical and protein analyses using proximity ligation assay, immunoblotting, mass cytometry and fluorescence-activated cell sorting, metabolomics, gene expression analysis, and real-time metabolic profiling utilizing the Seahorse XF analyzer. We used a Crohn's disease single-cell RNA sequencing dataset to infer the therapeutic relevance of targeting metabolic pathways in inflammatory Tregs. We examined the superior functionality of genetically modified Tregs in CD4+ T-cell-induced murine colitis models.
Results: Mitochondria-endoplasmic reticulum appositions, known to mediate pyruvate entry into mitochondria via voltage-dependent anion channel 1 (VDAC1), are abundant in Tregs. VDAC1 inhibition perturbed pyruvate metabolism, eliciting sensitization to other inflammatory signals reversible by membrane-permeable methyl pyruvate supplementation. Notably, interleukin (IL) 21 diminished mitochondria-endoplasmic reticulum appositions, resulting in enhanced enzymatic function of glycogen synthase kinase 3 β, a putative negative regulator of VDAC1, and a hypermetabolic state that amplified Treg inflammatory response. Methyl pyruvate and glycogen synthase kinase 3 β pharmacologic inhibitor (LY2090314) reversed IL21-induced metabolic rewiring and inflammatory state. Moreover, IL21-induced metabolic genes in Tregs in vitro were enriched in human Crohn's disease intestinal Tregs. Adoptively transferred Il21r-/- Tregs efficiently rescued murine colitis in contrast to wild-type Tregs.
Conclusions: IL21 triggers metabolic dysfunction associated with Treg inflammatory response. Inhibiting IL21-induced metabolism in Tregs may mitigate CD4+ T-cell-driven chronic intestinal inflammation.
Keywords: Inflammatory Bowel Disease; Interleukins; Mitochondria-ER Appositions; Pyruvate; Regulatory T Cells.
Copyright © 2024 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosures
The authors declare no conflicts of interest.
Figures


Update of
-
Interleukin-21 Drives a Hypermetabolic State and CD4+ T Cell-associated Pathogenicity in Chronic Intestinal Inflammation.bioRxiv [Preprint]. 2023 Jun 6:2023.06.02.543518. doi: 10.1101/2023.06.02.543518. bioRxiv. 2023. Update in: Gastroenterology. 2024 May;166(5):826-841.e19. doi: 10.1053/j.gastro.2024.01.026. PMID: 37333332 Free PMC article. Updated. Preprint.
References
-
- Friedrich M, Pohin M, Powrie F. Cytokine Networks in the Pathophysiology of Inflammatory Bowel Disease. Immunity 2019;50:992–1006. - PubMed
-
- Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2018;390:2769–2778. - PubMed
-
- Hirten RP, Iacucci M, Shah S, et al. Combining Biologics in Inflammatory Bowel Disease and Other Immune Mediated Inflammatory Disorders. Clin Gastroenterol Hepatol 2018;16:1374–1384. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

