Glutamate 139 of tropomyosin is critical for cardiac thin filament blocked-state stabilization
- PMID: 38266978
- PMCID: PMC11654406
- DOI: 10.1016/j.yjmcc.2024.01.004
Glutamate 139 of tropomyosin is critical for cardiac thin filament blocked-state stabilization
Abstract
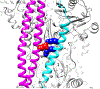
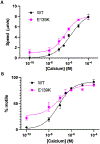
The cardiac thin filament proteins troponin and tropomyosin control actomyosin formation and thus cardiac contractility. Calcium binding to troponin changes tropomyosin position along the thin filament, allowing myosin head binding to actin required for heart muscle contraction. The thin filament regulatory proteins are hot spots for genetic mutations causing heart muscle dysfunction. While much of the thin filament structure has been characterized, critical regions of troponin and tropomyosin involved in triggering conformational changes remain unresolved. A poorly resolved region, helix-4 (H4) of troponin I, is thought to stabilize tropomyosin in a position on actin that blocks actomyosin interactions at low calcium concentrations during muscle relaxation. We have proposed that contact between glutamate 139 on tropomyosin and positively charged residues on H4 leads to blocking-state stabilization. In this study, we attempted to disrupt these interactions by replacing E139 with lysine (E139K) to define the importance of this residue in thin filament regulation. Comparison of mutant and wild-type tropomyosin was carried out using in-vitro motility assays, actin co-sedimentation, and molecular dynamics simulations to determine perturbations in troponin-tropomyosin function caused by the tropomyosin mutation. Motility assays revealed that mutant thin filaments moved at higher velocity at low calcium with increased calcium sensitivity demonstrating that tropomyosin residue 139 is vital for proper tropomyosin-mediated inhibition during relaxation. Similarly, molecular dynamic simulations revealed a mutation-induced decrease in interaction energy between tropomyosin-E139K and troponin I (R170 and K174). These results suggest that salt-bridge stabilization of tropomyosin position by troponin IH4 is essential to prevent actomyosin interactions during cardiac muscle relaxation.
Keywords: Blocked-state stabilization; Cardiac muscle regulation; Helix-4; In vitro motility; Tropomyosin; Troponin.
Copyright © 2024. Published by Elsevier Ltd.
Conflict of interest statement
Declaration of competing interest None.
Figures
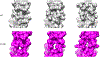
Similar articles
-
M8R tropomyosin mutation disrupts actin binding and filament regulation: The beginning affects the middle and end.J Biol Chem. 2020 Dec 11;295(50):17128-17137. doi: 10.1074/jbc.RA120.014713. Epub 2020 Oct 5. J Biol Chem. 2020. PMID: 33020181 Free PMC article.
-
HCM and DCM cardiomyopathy-linked α-tropomyosin mutations influence off-state stability and crossbridge interaction on thin filaments.Arch Biochem Biophys. 2018 Jun 1;647:84-92. doi: 10.1016/j.abb.2018.04.002. Epub 2018 Apr 5. Arch Biochem Biophys. 2018. PMID: 29626422 Free PMC article.
-
C-terminal troponin-I residues trap tropomyosin in the muscle thin filament blocked-state.Biochem Biophys Res Commun. 2021 Apr 30;551:27-32. doi: 10.1016/j.bbrc.2021.03.010. Epub 2021 Mar 11. Biochem Biophys Res Commun. 2021. PMID: 33714756 Free PMC article.
-
Regulation of contraction in striated muscle.Physiol Rev. 2000 Apr;80(2):853-924. doi: 10.1152/physrev.2000.80.2.853. Physiol Rev. 2000. PMID: 10747208 Review.
-
Troponin Revealed: Uncovering the Structure of the Thin Filament On-Off Switch in Striated Muscle.Biophys J. 2021 Jan 5;120(1):1-9. doi: 10.1016/j.bpj.2020.11.014. Epub 2020 Nov 20. Biophys J. 2021. PMID: 33221250 Free PMC article. Review.
Cited by
-
Dual role of Tropomyosin-R160 in thin filament regulation: Insights into phosphorylation-dependent cardiac relaxation and cardiomyopathy mechanisms.Arch Biochem Biophys. 2025 Jun;768:110380. doi: 10.1016/j.abb.2025.110380. Epub 2025 Mar 6. Arch Biochem Biophys. 2025. PMID: 40057222
-
Modeling the effects of thin filament near-neighbor cooperative interactions in mammalian myocardium.J Gen Physiol. 2025 Mar 3;157(2):e202413582. doi: 10.1085/jgp.202413582. Epub 2025 Jan 27. J Gen Physiol. 2025. PMID: 39869069 Free PMC article.
-
Protein Sequencing with Single Amino Acid Resolution Discerns Peptides That Discriminate Tropomyosin Proteoforms.J Proteome Res. 2025 Aug 1;24(8):3798-3807. doi: 10.1021/acs.jproteome.4c00978. Epub 2025 Jun 10. J Proteome Res. 2025. PMID: 40493858 Free PMC article.
-
Velcro-binding by cardiac troponin-I traps tropomyosin on actin in a low-energy relaxed state.Biochem Biophys Res Commun. 2025 Apr 9;757:151595. doi: 10.1016/j.bbrc.2025.151595. Epub 2025 Mar 8. Biochem Biophys Res Commun. 2025. PMID: 40088678
-
Generation and Accumulation of Various Advanced Glycation End-Products in Cardiomyocytes May Induce Cardiovascular Disease.Int J Mol Sci. 2024 Jul 3;25(13):7319. doi: 10.3390/ijms25137319. Int J Mol Sci. 2024. PMID: 39000424 Free PMC article. Review.
References
-
- Huxley AF, Niedergerke R, Structural changes in muscle during contraction: interference microscopy of living muscle Fibres, Nature 173 (1954) 971–973. - PubMed
-
- Huxley H, Hanson J, Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation, Nature 173 (4412) (1954) 973–976. - PubMed
-
- Robaszkiewicz K, et al., Impaired tropomyosin-troponin interactions reduce activation of the actin thin filament, Biochim. Biophys. Acta 1854 (5) (2015) 381–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

