Anti-correlated feature selection prevents false discovery of subpopulations in scRNAseq
- PMID: 38267438
- PMCID: PMC10808220
- DOI: 10.1038/s41467-023-43406-9
Anti-correlated feature selection prevents false discovery of subpopulations in scRNAseq
Abstract
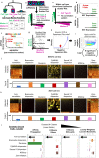
While sub-clustering cell-populations has become popular in single cell-omics, negative controls for this process are lacking. Popular feature-selection/clustering algorithms fail the null-dataset problem, allowing erroneous subdivisions of homogenous clusters until nearly each cell is called its own cluster. Using real and synthetic datasets, we find that anti-correlated gene selection reduces or eliminates erroneous subdivisions, increases marker-gene selection efficacy, and efficiently scales to millions of cells.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

