Motion of VAPB molecules reveals ER-mitochondria contact site subdomains
- PMID: 38267577
- PMCID: PMC10830423
- DOI: 10.1038/s41586-023-06956-y
Motion of VAPB molecules reveals ER-mitochondria contact site subdomains
Abstract
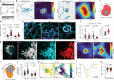
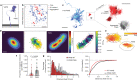
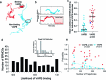
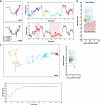
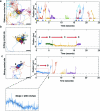
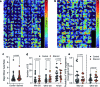
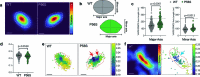
To coordinate cellular physiology, eukaryotic cells rely on the rapid exchange of molecules at specialized organelle-organelle contact sites1,2. Endoplasmic reticulum-mitochondrial contact sites (ERMCSs) are particularly vital communication hubs, playing key roles in the exchange of signalling molecules, lipids and metabolites3,4. ERMCSs are maintained by interactions between complementary tethering molecules on the surface of each organelle5,6. However, due to the extreme sensitivity of these membrane interfaces to experimental perturbation7,8, a clear understanding of their nanoscale organization and regulation is still lacking. Here we combine three-dimensional electron microscopy with high-speed molecular tracking of a model organelle tether, Vesicle-associated membrane protein (VAMP)-associated protein B (VAPB), to map the structure and diffusion landscape of ERMCSs. We uncovered dynamic subdomains within VAPB contact sites that correlate with ER membrane curvature and undergo rapid remodelling. We show that VAPB molecules enter and leave ERMCSs within seconds, despite the contact site itself remaining stable over much longer time scales. This metastability allows ERMCSs to remodel with changes in the physiological environment to accommodate metabolic needs of the cell. An amyotrophic lateral sclerosis-associated mutation in VAPB perturbs these subdomains, likely impairing their remodelling capacity and resulting in impaired interorganelle communication. These results establish high-speed single-molecule imaging as a new tool for mapping the structure of contact site interfaces and reveal that the diffusion landscape of VAPB at contact sites is a crucial component of ERMCS homeostasis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: C.P.C. is the founder and employed by Ursa Analytics, a company that offers statistical analysis services such as the non-parametric Bayesian approach utilized in this paper. All other authors declare no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

