Disulfiram ameliorates STING/MITA-dependent inflammation and autoimmunity by targeting RNF115
- PMID: 38267694
- PMCID: PMC10901794
- DOI: 10.1038/s41423-024-01131-3
Disulfiram ameliorates STING/MITA-dependent inflammation and autoimmunity by targeting RNF115
Abstract
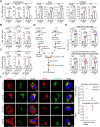
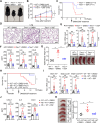
STING (also known as MITA) is an adaptor protein that mediates cytoplasmic DNA-triggered signaling, and aberrant activation of STING/MITA by cytosolic self-DNA or gain-of-function mutations causes severe inflammation. Here, we show that STING-mediated inflammation and autoimmunity are promoted by RNF115 and alleviated by the RNF115 inhibitor disulfiram (DSF). Knockout of RNF115 or treatment with DSF significantly inhibit systemic inflammation and autoimmune lethality and restore immune cell development in Trex1-/- mice and STINGN153S/WT bone marrow chimeric mice. In addition, knockdown or pharmacological inhibition of RNF115 substantially downregulate the expression of IFN-α, IFN-γ and proinflammatory cytokines in PBMCs from patients with systemic lupus erythematosus (SLE) who exhibit high concentrations of dsDNA in peripheral blood. Mechanistically, knockout or inhibition of RNF115 impair the oligomerization and Golgi localization of STING in various types of cells transfected with cGAMP and in organs and cells from Trex1-/- mice. Interestingly, knockout of RNF115 inhibits the activation and Golgi localization of STINGN153S as well as the expression of proinflammatory cytokines in myeloid cells but not in endothelial cells or fibroblasts. Taken together, these findings highlight the RNF115-mediated cell type-specific regulation of STING and STINGN153S and provide potential targeted intervention strategies for STING-related autoimmune diseases.
Keywords: Autoimmunity; Disulfiram; RNF115; SLE; STING/MITA.
© 2024. The Author(s), under exclusive licence to CSI and USTC.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

