Increasing the accuracy of single-molecule data analysis using tMAVEN
- PMID: 38268189
- PMCID: PMC11393709
- DOI: 10.1016/j.bpj.2024.01.022
Increasing the accuracy of single-molecule data analysis using tMAVEN
Abstract
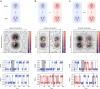
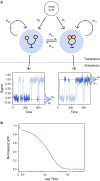
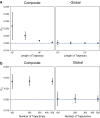
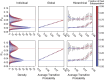
Time-dependent single-molecule experiments contain rich kinetic information about the functional dynamics of biomolecules. A key step in extracting this information is the application of kinetic models, such as hidden Markov models (HMMs), which characterize the molecular mechanism governing the experimental system. Unfortunately, researchers rarely know the physicochemical details of this molecular mechanism a priori, which raises questions about how to select the most appropriate kinetic model for a given single-molecule data set and what consequences arise if the wrong model is chosen. To address these questions, we have developed and used time-series modeling, analysis, and visualization environment (tMAVEN), a comprehensive, open-source, and extensible software platform. tMAVEN can perform each step of the single-molecule analysis pipeline, from preprocessing to kinetic modeling to plotting, and has been designed to enable the analysis of a single-molecule data set with multiple types of kinetic models. Using tMAVEN, we have systematically investigated mismatches between kinetic models and molecular mechanisms by analyzing simulated examples of prototypical single-molecule data sets exhibiting common experimental complications, such as molecular heterogeneity, with a series of different types of HMMs. Our results show that no single kinetic modeling strategy is mathematically appropriate for all experimental contexts. Indeed, HMMs only correctly capture the underlying molecular mechanism in the simplest of cases. As such, researchers must modify HMMs using physicochemical principles to avoid the risk of missing the significant biological and biophysical insights into molecular heterogeneity that their experiments provide. By enabling the facile, side-by-side application of multiple types of kinetic models to individual single-molecule data sets, tMAVEN allows researchers to carefully tailor their modeling approach to match the complexity of the underlying biomolecular dynamics and increase the accuracy of their single-molecule data analyses.
Copyright © 2024 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Update of
-
Increasing the accuracy of single-molecule data analysis using tMAVEN.bioRxiv [Preprint]. 2024 Jan 21:2023.08.15.553409. doi: 10.1101/2023.08.15.553409. bioRxiv. 2024. Update in: Biophys J. 2024 Sep 3;123(17):2765-2780. doi: 10.1016/j.bpj.2024.01.022. PMID: 37645812 Free PMC article. Updated. Preprint.
References
-
- Bustamante C., Bryant Z., Smith S.B. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. - PubMed
-
- MacDougall D.D., Fei J., Gonzalez R.L. In: Molecular Machines in Biology. Frank J., editor. Cambridge University Press; Cambridge: 2011. Single-Molecule Fluorescence Resonance Energy Transfer Investigations of Ribosome-Catalyzed Protein Synthesis; pp. 93–116.
-
- Du C., Kou S.C. Statistical Methodology in Single-Molecule Experiments. Stat. Sci. 2020;35:75–91.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

