Microfluidics enabled multi-omics triple-shot mass spectrometry for cell-based therapies
- PMID: 38268742
- PMCID: PMC10807926
- DOI: 10.1063/5.0175178
Microfluidics enabled multi-omics triple-shot mass spectrometry for cell-based therapies
Abstract
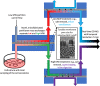
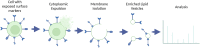
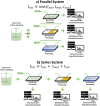
In recent years, cell-based therapies have transformed medical treatment. These therapies present a multitude of challenges associated with identifying the mechanism of action, developing accurate safety and potency assays, and achieving low-cost product manufacturing at scale. The complexity of the problem can be attributed to the intricate composition of the therapeutic products: living cells with complex biochemical compositions. Identifying and measuring critical quality attributes (CQAs) that impact therapy success is crucial for both the therapy development and its manufacturing. Unfortunately, current analytical methods and tools for identifying and measuring CQAs are limited in both scope and speed. This Perspective explores the potential for microfluidic-enabled mass spectrometry (MS) systems to comprehensively characterize CQAs for cell-based therapies, focusing on secretome, intracellular metabolome, and surfaceome biomarkers. Powerful microfluidic sampling and processing platforms have been recently presented for the secretome and intracellular metabolome, which could be implemented with MS for fast, locally sampled screening of the cell culture. However, surfaceome analysis remains limited by the lack of rapid isolation and enrichment methods. Developing innovative microfluidic approaches for surface marker analysis and integrating them with secretome and metabolome measurements using a common analytical platform hold the promise of enhancing our understanding of CQAs across all "omes," potentially revolutionizing cell-based therapy development and manufacturing for improved efficacy and patient accessibility.
© 2024 Author(s).
Conflict of interest statement
A.G.F., M.A.C., and A.L.C. are pursuing commercialization of the DSP technology discussed in this article. The terms of this arrangement have been reviewed and approved by Georgia Tech in accordance with its conflict-of-interest policies. The remaining authors declare no competing interests.
Figures


References
-
- Bordignon C., Carlo-Stella C., Colombo M. P., De Vincentiis A., Lanata L., Lemoli R. M., Locatelli F., Olivieri A., Rondelli D., Zanon P., and Tura S., Haematologica 84(12), 1110–1149 (1999), https://www.webofscience.com/wos/woscc/full-record/WOS:000084089700012?S.... - PubMed
-
- “Approved cellular and gene therapy products” [Office of Tissues and Advanced Therapies (OATA), 2023], https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-produ....
Grants and funding
LinkOut - more resources
Full Text Sources
