Statistical framework to determine indel-length distribution
- PMID: 38269647
- PMCID: PMC10868340
- DOI: 10.1093/bioinformatics/btae043
Statistical framework to determine indel-length distribution
Abstract
Motivation: Insertions and deletions (indels) of short DNA segments, along with substitutions, are the most frequent molecular evolutionary events. Indels were shown to affect numerous macro-evolutionary processes. Because indels may span multiple positions, their impact is a product of both their rate and their length distribution. An accurate inference of indel-length distribution is important for multiple evolutionary and bioinformatics applications, most notably for alignment software. Previous studies counted the number of continuous gap characters in alignments to determine the best-fitting length distribution. However, gap-counting methods are not statistically rigorous, as gap blocks are not synonymous with indels. Furthermore, such methods rely on alignments that regularly contain errors and are biased due to the assumption of alignment methods that indels lengths follow a geometric distribution.
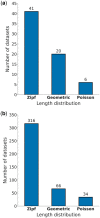
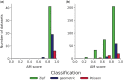
Results: We aimed to determine which indel-length distribution best characterizes alignments using statistical rigorous methodologies. To this end, we reduced the alignment bias using a machine-learning algorithm and applied an Approximate Bayesian Computation methodology for model selection. Moreover, we developed a novel method to test if current indel models provide an adequate representation of the evolutionary process. We found that the best-fitting model varies among alignments, with a Zipf length distribution fitting the vast majority of them.
Availability and implementation: The data underlying this article are available in Github, at https://github.com/elyawy/SpartaSim and https://github.com/elyawy/SpartaPipeline.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

