CLPB disaggregase dysfunction impacts the functional integrity of the proteolytic SPY complex
- PMID: 38270563
- PMCID: PMC10818064
- DOI: 10.1083/jcb.202305087
CLPB disaggregase dysfunction impacts the functional integrity of the proteolytic SPY complex
Abstract
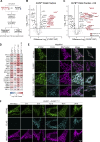
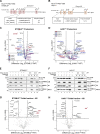
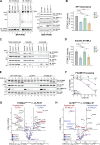
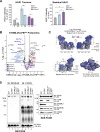
CLPB is a mitochondrial intermembrane space AAA+ domain-containing disaggregase. CLPB mutations are associated with 3-methylglutaconic aciduria and neutropenia; however, the molecular mechanism underscoring disease and the contribution of CLPB substrates to disease pathology remains unknown. Interactions between CLPB and mitochondrial quality control (QC) factors, including PARL and OPA1, have been reported, hinting at dysregulation of organelle QC in disease. Utilizing proteomic and biochemical approaches, we show a stress-specific aggregation phenotype in a CLPB-null environment and define the CLPB substrate profile. We illustrate an interplay between intermembrane space proteins including CLPB, HAX1, HTRA2, and the inner membrane quality control proteins (STOML2, PARL, YME1L1; SPY complex), with CLPB deficiency impeding SPY complex function by virtue of protein aggregation in the intermembrane space. We conclude that there is an interdependency of mitochondrial QC components at the intermembrane space/inner membrane interface, and perturbations to this network may underscore CLPB disease pathology.
© 2024 Baker et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

