Invasive growth of brain metastases is linked to CHI3L1 release from pSTAT3-positive astrocytes
- PMID: 38271182
- PMCID: PMC11145453
- DOI: 10.1093/neuonc/noae013
Invasive growth of brain metastases is linked to CHI3L1 release from pSTAT3-positive astrocytes
Abstract
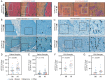
Background: Compared to minimally invasive brain metastases (MI BrM), highly invasive (HI) lesions form abundant contacts with cells in the peritumoral brain parenchyma and are associated with poor prognosis. Reactive astrocytes (RAs) labeled by phosphorylated STAT3 (pSTAT3) have recently emerged as a promising therapeutic target for BrM. Here, we explore whether the BrM invasion pattern is influenced by pSTAT3+ RAs and may serve as a predictive biomarker for STAT3 inhibition.
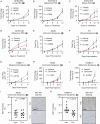
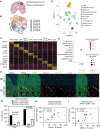
Methods: We used immunohistochemistry to identify pSTAT3+ RAs in HI and MI human and patient-derived xenograft (PDX) BrM. Using PDX, syngeneic, and transgenic mouse models of HI and MI BrM, we assessed how pharmacological STAT3 inhibition or RA-specific STAT3 genetic ablation affected BrM growth in vivo. Cancer cell invasion was modeled in vitro using a brain slice-tumor co-culture assay. We performed single-cell RNA sequencing of human BrM and adjacent brain tissue.
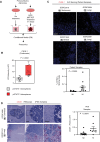
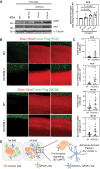
Results: RAs expressing pSTAT3 are situated at the brain-tumor interface and drive BrM invasive growth. HI BrM invasion pattern was associated with delayed growth in the context of STAT3 inhibition or genetic ablation. We demonstrate that pSTAT3+ RAs secrete Chitinase 3-like-1 (CHI3L1), which is a known STAT3 transcriptional target. Furthermore, single-cell RNA sequencing identified CHI3L1-expressing RAs in human HI BrM. STAT3 activation, or recombinant CHI3L1 alone, induced cancer cell invasion into the brain parenchyma using a brain slice-tumor plug co-culture assay.
Conclusions: Together, these data reveal that pSTAT3+ RA-derived CHI3L1 is associated with BrM invasion, implicating STAT3 and CHI3L1 as clinically relevant therapeutic targets for the treatment of HI BrM.
Keywords: CHI3L1; STAT3; astrocyte; brain metastasis; histopathological growth patterns; invasion.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures

References
-
- Suh JH, Kotecha R, Chao ST, et al.. Current approaches to the management of brain metastases. Nat Rev Clin Oncol. 2020;17(5):279–299. - PubMed
-
- Nayak L, Lee EQ, Wen PY.. Epidemiology of brain metastases. Curr Oncol Rep. 2012;14(1):48–54. - PubMed
-
- Singh Achrol A, Rennert RC, Anders C, et al.. Brain metastases. Nat Rev Dis Prim. 2019;5(5):1–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

