ELONGATED HYPOCOTYL 5a modulates FLOWERING LOCUS T2 and gibberellin levels to control dormancy and bud break in poplar
- PMID: 38271284
- PMCID: PMC11062467
- DOI: 10.1093/plcell/koae022
ELONGATED HYPOCOTYL 5a modulates FLOWERING LOCUS T2 and gibberellin levels to control dormancy and bud break in poplar
Abstract
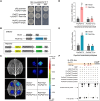
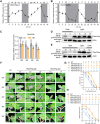
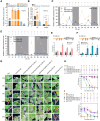
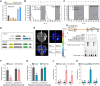
Photoperiod is a crucial environmental cue for phenological responses, including growth cessation and winter dormancy in perennial woody plants. Two regulatory modules within the photoperiod pathway explain bud dormancy induction in poplar (Populus spp.): the circadian oscillator LATE ELONGATED HYPOCOTYL 2 (LHY2) and GIGANTEA-like genes (GIs) both regulate the key target for winter dormancy induction FLOWERING LOCUS T2 (FT2). However, modification of LHY2 and GIs cannot completely prevent growth cessation and bud set under short-day (SD) conditions, indicating that additional regulatory modules are likely involved. We identified PtoHY5a, an orthologs of the photomorphogenesis regulatory factor ELONGATED HYPOCOTYL 5 (HY5) in poplar (Populus tomentosa), that directly activates PtoFT2 expression and represses the circadian oscillation of LHY2, indirectly activating PtoFT2 expression. Thus, PtoHY5a suppresses SD-induced growth cessation and bud set. Accordingly, PtoHY5a knockout facilitates dormancy induction. PtoHY5a also inhibits bud-break in poplar by controlling gibberellic acid (GA) levels in apical buds. Additionally, PtoHY5a regulates the photoperiodic control of seasonal growth downstream of phytochrome PHYB2. Thus, PtoHY5a modulates seasonal growth in poplar by regulating the PtoPHYB2-PtoHY5a-PtoFT2 module to determine the onset of winter dormancy, and by fine-tuning GA levels to control bud-break.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

