Airway "Resistotypes" and Clinical Outcomes in Bronchiectasis
- PMID: 38271608
- PMCID: PMC11197066
- DOI: 10.1164/rccm.202306-1059OC
Airway "Resistotypes" and Clinical Outcomes in Bronchiectasis
Abstract
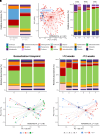
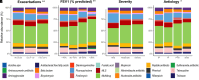
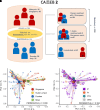
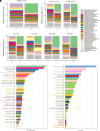
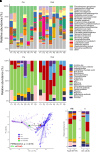
Rationale: Chronic infection and inflammation shapes the airway microbiome in bronchiectasis. Utilizing whole-genome shotgun metagenomics to analyze the airway resistome provides insight into interplay between microbes, resistance genes, and clinical outcomes. Objectives: To apply whole-genome shotgun metagenomics to the airway microbiome in bronchiectasis to highlight a diverse pool of antimicrobial resistance genes: the "resistome," the clinical significance of which remains unclear. Methods: Individuals with bronchiectasis were prospectively recruited into cross-sectional and longitudinal cohorts (n = 280), including the international multicenter cross-sectional Cohort of Asian and Matched European Bronchiectasis 2 (CAMEB 2) study (n = 251) and two independent cohorts, one describing patients experiencing acute exacerbation and a further cohort of patients undergoing Pseudomonas aeruginosa eradication treatment. Sputum was subjected to metagenomic sequencing, and the bronchiectasis resistome was evaluated in association with clinical outcomes and underlying host microbiomes. Measurements and Main Results: The bronchiectasis resistome features a unique resistance gene profile and increased counts of aminoglycoside, bicyclomycin, phenicol, triclosan, and multidrug resistance genes. Longitudinally, it exhibits within-patient stability over time and during exacerbations despite between-patient heterogeneity. Proportional differences in baseline resistome profiles, including increased macrolide and multidrug resistance genes, associate with shorter intervals to the next exacerbation, whereas distinct resistome archetypes associate with frequent exacerbations, poorer lung function, geographic origin, and the host microbiome. Unsupervised analysis of resistome profiles identified two clinically relevant "resistotypes," RT1 and RT2, the latter characterized by poor clinical outcomes, increased multidrug resistance, and P. aeruginosa. Successful targeted eradication in P. aeruginosa-colonized individuals mediated reversion from RT2 to RT1, a more clinically favorable resistome profile demonstrating reduced resistance gene diversity. Conclusions: The bronchiectasis resistome associates with clinical outcomes, geographic origin, and the underlying host microbiome. Bronchiectasis resistotypes link to clinical disease and are modifiable through targeted antimicrobial therapy.
Keywords: bronchiectasis; metagenomics; microbiome; resistome; resistotype.
Figures

Comment in
-
Vive la Resistome: Are We Ready for a Metagenomics Revolution in Bronchiectasis?Am J Respir Crit Care Med. 2024 Jul 1;210(1):10-12. doi: 10.1164/rccm.202402-0352ED. Am J Respir Crit Care Med. 2024. PMID: 38530113 Free PMC article. No abstract available.
References
-
- Mac Aogáin M, Chotirmall SH. Microbiology and the microbiome in bronchiectasis. Clin Chest Med . 2022;43:23–34. - PubMed
-
- Budden KF, Shukla SD, Rehman SF, Bowerman KL, Keely S, Hugenholtz P, et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir Med . 2019;7:907–920. - PubMed
-
- Chotirmall SH, Chalmers JD. RESPIRE: breathing new life into bronchiectasis. Eur Respir J . 2018;51:1702444. - PubMed

