Genetic variants affecting NQO1 protein levels impact the efficacy of idebenone treatment in Leber hereditary optic neuropathy
- PMID: 38272025
- PMCID: PMC10897523
- DOI: 10.1016/j.xcrm.2023.101383
Genetic variants affecting NQO1 protein levels impact the efficacy of idebenone treatment in Leber hereditary optic neuropathy
Abstract
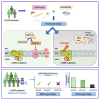
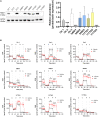
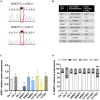
Idebenone, the only approved treatment for Leber hereditary optic neuropathy (LHON), promotes recovery of visual function in up to 50% of patients, but we can neither predict nor understand the non-responders. Idebenone is reduced by the cytosolic NAD(P)H oxidoreductase I (NQO1) and directly shuttles electrons to respiratory complex III, bypassing complex I affected in LHON. We show here that two polymorphic variants drastically reduce NQO1 protein levels when homozygous or compound heterozygous. This hampers idebenone reduction. In its oxidized form, idebenone inhibits complex I, decreasing respiratory function in cells. By retrospectively analyzing a large cohort of idebenone-treated LHON patients, classified by their response to therapy, we show that patients with homozygous or compound heterozygous NQO1 variants have the poorest therapy response, particularly if carrying the m.3460G>A/MT-ND1 LHON mutation. These results suggest consideration of patient NQO1 genotype and mitochondrial DNA mutation in the context of idebenone therapy.
Keywords: LHON; Leber hereditary optic neuropathy; NQO1; complex I; cybrids; fibroblasts; idebenone; mtDNA; retinal ganglion cells.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.R., P.B., M. Carbonelli, G.A., C.L.M., C.B.C., T.K., and V.C. are involved in clinical trials with idebenone (Santhera Pharmaceuticals and Chiesi Farmaceutici) in LHON patients. M.R., A.A.S., P.B., M. Carbonelli, G.A., C.L.M., C.B.C., T.K., and V.C. are involved in gene therapy trials with Lumevoq (GenSight Biologics) in LHON patients. A.A.S., C.L.M., T.K., and V.C. have received research support, speaker honoraria, consulting fees, and travel reimbursement from Santhera Pharmaceuticals, Chiesi GmbH, and GenSight Biologics. None of these activities are related to conduct of this study or writing of the manuscript.
Figures





References
-
- Zs -Nagy I. Chemistry, toxicology, pharmacology and pharmacokinetics of idebenone: a review. Arch. Gerontol. Geriatr. 1990;11:177–186. - PubMed
-
- Gillis J.C., Benefield P., McTavish D. Idebenone: A Review of its Pharmacodynamic and Pharmacokinetic Properties, and Therapeutic Use in Age-Related Cognitive Disorders. Drugs Aging. 1994;5:133–152. - PubMed
-
- Varricchio C., Beirne K., Heard C., Newland B., Rozanowska M., Brancale A., Votruba M. The ying and yang of idebenone: Not too little, not too much – cell death in NQO1 deficient cells and the mouse retina. Free Radic. Biol. Med. 2020;152:551–560. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

