Semi-automated sequence curation for reliable reference datasets in ITS2 vascular plant DNA (meta-)barcoding
- PMID: 38272945
- PMCID: PMC10810873
- DOI: 10.1038/s41597-024-02962-5
Semi-automated sequence curation for reliable reference datasets in ITS2 vascular plant DNA (meta-)barcoding
Abstract
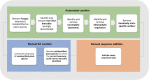
One of the most critical steps for accurate taxonomic identification in DNA (meta)-barcoding is to have an accurate DNA reference sequence dataset for the marker of choice. Therefore, developing such a dataset has been a long-term ambition, especially in the Viridiplantae kingdom. Typically, reference datasets are constructed with sequences downloaded from general public databases, which can carry taxonomic and other relevant errors. Herein, we constructed a curated (i) global dataset, (ii) European crop dataset, and (iii) 27 datasets for the EU countries for the ITS2 barcoding marker of vascular plants. To that end, we first developed a pipeline script that entails (i) an automated curation stage comprising five filters, (ii) manual taxonomic correction for misclassified taxa, and (iii) manual addition of newly sequenced species. The pipeline allows easy updating of the curated datasets. With this approach, 13% of the sequences, corresponding to 7% of species originally imported from GenBank, were discarded. Further, 259 sequences were manually added to the curated global dataset, which now comprises 307,977 sequences of 111,382 plant species.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

