Computationally designed sensors detect endogenous Ras activity and signaling effectors at subcellular resolution
- PMID: 38273065
- PMCID: PMC11631767
- DOI: 10.1038/s41587-023-02107-w
Computationally designed sensors detect endogenous Ras activity and signaling effectors at subcellular resolution
Abstract
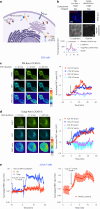
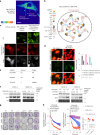
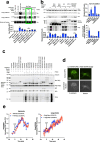
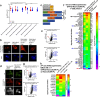
The utility of genetically encoded biosensors for sensing the activity of signaling proteins has been hampered by a lack of strategies for matching sensor sensitivity to the physiological concentration range of the target. Here we used computational protein design to generate intracellular sensors of Ras activity (LOCKR-based Sensor for Ras activity (Ras-LOCKR-S)) and proximity labelers of the Ras signaling environment (LOCKR-based, Ras activity-dependent Proximity Labeler (Ras-LOCKR-PL)). These tools allow the detection of endogenous Ras activity and labeling of the surrounding environment at subcellular resolution. Using these sensors in human cancer cell lines, we identified Ras-interacting proteins in oncogenic EML4-Alk granules and found that Src-Associated in Mitosis 68-kDa (SAM68) protein specifically enhances Ras activity in the granules. The ability to subcellularly localize endogenous Ras activity should deepen our understanding of Ras function in health and disease and may suggest potential therapeutic strategies.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: J.Z.Z., D.J.M. and D.B. are co-inventors on a provisional patent application (application number 63/380,884, submitted by the University of Washington) covering the biosensors described in this paper. The other authors declare no competing interests.
Figures





References
-
- Bivona, T. G. et al. PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol. Cell21, 481–493 (2006). - PubMed
-
- Chiu, V. K. et al. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol.4, 343–350 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

