Mitochondrial CISD1/Cisd accumulation blocks mitophagy and genetic or pharmacological inhibition rescues neurodegenerative phenotypes in Pink1/parkin models
- PMID: 38273330
- PMCID: PMC10811860
- DOI: 10.1186/s13024-024-00701-3
Mitochondrial CISD1/Cisd accumulation blocks mitophagy and genetic or pharmacological inhibition rescues neurodegenerative phenotypes in Pink1/parkin models
Abstract
Background: Mitochondrial dysfunction and toxic protein aggregates have been shown to be key features in the pathogenesis of neurodegenerative diseases, such as Parkinson's disease (PD). Functional analysis of genes linked to PD have revealed that the E3 ligase Parkin and the mitochondrial kinase PINK1 are important factors for mitochondrial quality control. PINK1 phosphorylates and activates Parkin, which in turn ubiquitinates mitochondrial proteins priming them and the mitochondrion itself for degradation. However, it is unclear whether dysregulated mitochondrial degradation or the toxic build-up of certain Parkin ubiquitin substrates is the driving pathophysiological mechanism leading to PD. The iron-sulphur cluster containing proteins CISD1 and CISD2 have been identified as major targets of Parkin in various proteomic studies.
Methods: We employed in vivo Drosophila and human cell culture models to study the role of CISD proteins in cell and tissue viability as well as aged-related neurodegeneration, specifically analysing aspects of mitophagy and autophagy using orthogonal assays.
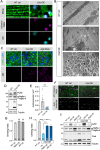
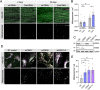
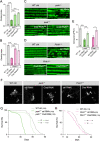
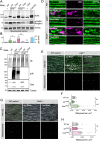
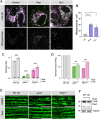
Results: We show that the Drosophila homolog Cisd accumulates in Pink1 and parkin mutant flies, as well as during ageing. We observed that build-up of Cisd is particularly toxic in neurons, resulting in mitochondrial defects and Ser65-phospho-Ubiquitin accumulation. Age-related increase of Cisd blocks mitophagy and impairs autophagy flux. Importantly, reduction of Cisd levels upregulates mitophagy in vitro and in vivo, and ameliorates pathological phenotypes in locomotion, lifespan and neurodegeneration in Pink1/parkin mutant flies. In addition, we show that pharmacological inhibition of CISD1/2 by rosiglitazone and NL-1 induces mitophagy in human cells and ameliorates the defective phenotypes of Pink1/parkin mutants.
Conclusion: Altogether, our studies indicate that Cisd accumulation during ageing and in Pink1/parkin mutants is a key driver of pathology by blocking mitophagy, and genetically and pharmacologically inhibiting CISD proteins may offer a potential target for therapeutic intervention.
Keywords: Ageing; Autophagy; CISD1; CISD2; Cisd; Mitochondria; Mitophagy; Neurodegeneration; PINK1; Parkin; Parkinson’s disease.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, et al. Parkinson disease. Nat Rev Dis Primer. 2017;3:1–21. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

