Seasonal and spatial variations of Synechococcus in abundance, pigment types, and genetic diversity in a temperate semi-enclosed bay
- PMID: 38274747
- PMCID: PMC10808157
- DOI: 10.3389/fmicb.2023.1322548
Seasonal and spatial variations of Synechococcus in abundance, pigment types, and genetic diversity in a temperate semi-enclosed bay
Abstract
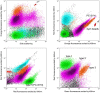
Synechococcus is abundant and globally widespread in various marine environments. Seasonal and spatial variations in Synechococcus abundance, pigment types, and genetic diversity were investigated based on flow cytometric analysis and high-throughput sequencing of cpcBA operon (encoding phycocyanin) and rpoC1 gene (encoding RNA polymerase) in a temperate semi-enclosed bay. Synechococcus abundance exhibited seasonal variations with the highest value in summer and the lowest value in winter, which was consistent with temperature variation. Three pigment types of Synechococcus type 1, type 2, and type 3 were distinguished based on cpcBA operon, which displayed obvious variations spatially between the inner and the outer bay. Freshwater discharge and water turbidity played important roles in regulating Synechococcus pigment types. Synechococcus assemblages were phylogenetically diverse (12 different lineages) based on rpoC1 gene and dominated by three core lineages S5.1-I, S5.1-IX, and S5.2-CB5 in different seasons. Our study demonstrated that Synechococcus abundance, pigment types, and genetic diversity displayed variations seasonally and spatially by different techniques, which were mainly driven by temperature, salinity, nutrients, and turbidity. The combination of more technical means provides more information for studying Synechococcus distribution. In this study, three pigment types of Synechococcus were discriminated simultaneously by dual lasers flow cytometer for the first time.
Keywords: Synechococcus; co-dominate; cpcBA operon; genetic diversity; pigment types; rpoC1 gene; temperate semi-enclosed bay.
Copyright © 2024 Li, Dong, Sun, Zhao, Zhao, Zhang and Xiao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Agawin N. S. R., Duarte C. M., Agusti S. (1998). Growth and abundance of Synechococcus sp. in a Mediterranean Bay: seasonality and relationship with temperature. Mar. Ecol.-Prog. Ser. 170, 45–53. doi: 10.3354/meps170045 - DOI
-
- Chen F., Wang K., Kan J., Bachoon D., Lu J. (2004). Phylogenetic diversity of Synechococcus in the Chesapeake Bay revealed by Ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO) large subunit gene (rbcL) sequences. Aquat. Microb. Ecol. 36, 153–164. doi: 10.3354/ame036153 - DOI
LinkOut - more resources
Full Text Sources

