DEK deficiency suppresses mitophagy to protect against house dust mite-induced asthma
- PMID: 38274803
- PMCID: PMC10808738
- DOI: 10.3389/fimmu.2023.1289774
DEK deficiency suppresses mitophagy to protect against house dust mite-induced asthma
Abstract
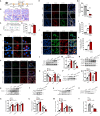
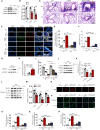
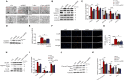
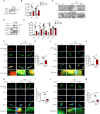
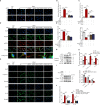
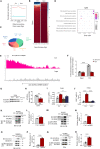
DEK protein is highly expressed in asthma. However, the mechanism of DEK on mitophagy in asthma has not been fully understood. This study aims to investigate the role and mechanism of DEK in asthmatic airway inflammation and in regulating PINK1-Parkin-mediated mitophagy, NLRP3 inflammasome activation, and apoptosis. PINK1-Parkin mitophagy, NLRP3 inflammasome, and apoptosis were examined after gene silencing or treatment with specific inhibitors (MitoTEMPO, MCC950, and Ac-DEVD-CHO) in house dust mite (HDM) or recombinant DEK (rmDEK)-induced WT and DEK-/- asthmatic mice and BEAS-2B cells. The regulatory role of DEK on ATAD3A was detected using ChIP-sequence and co-immunoprecipitation. rmDEK promoted eosinophil recruitment, and co-localization of TOM20 and LC3B, MFN1 and mitochondria, LC3B and VDAC, and ROS generation, reduced protein level of MnSOD in HDM induced-asthmatic mice. Moreover, rmDEK also increased DRP1 expression, PINK1-Parkin-mediated mitophagy, NLRP3 inflammasome activation, and apoptosis. These effects were partially reversed in DEK-/- mice. In BEAS-2B cells, siDEK diminished the Parkin, LC3B, and DRP1 translocation to mitochondria, mtROS, TOM20, and mtDNA. ChIP-sequence analysis showed that DEK was enriched on the ATAD3A promoter and could positively regulate ATAD3A expression. Additionally, ATAD3A was highly expressed in HDM-induced asthma models and interacted with DRP1, and siATAD3A could down-regulate DRP1 and mtDNA-mediated mitochondrial oxidative damage. Conclusively, DEK deficiency alleviates airway inflammation in asthma by down-regulating PINK1-Parkin mitophagy, NLRP3 inflammasome activation, and apoptosis. The mechanism may be through the DEK/ATAD3A/DRP1 signaling axis. Our findings may provide new potential therapeutic targets for asthma treatment.
Keywords: DEK; NLRP3 inflammasome; PINK1-Parkin; asthma; mitophagy.
Copyright © 2024 Bai, Liu, Quan, Han, Wang, Wang, Wang, Li, Li, Piao, Song and Yan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Comment on
-
Parkin, an E3 ubiquitin ligase, enhances airway mitochondrial DNA release and inflammation.Thorax. 2020 Sep;75(9):717-724. doi: 10.1136/thoraxjnl-2019-214158. Epub 2020 Jun 4. Thorax. 2020. PMID: 32499407
Similar articles
-
Polydatin inhibits mitochondrial damage and mitochondrial ROS by promoting PINK1-Parkin-mediated mitophagy in allergic rhinitis.FASEB J. 2023 Apr;37(4):e22852. doi: 10.1096/fj.202201231RR. FASEB J. 2023. PMID: 36906289
-
PINK1-parkin pathway of mitophagy protects against contrast-induced acute kidney injury via decreasing mitochondrial ROS and NLRP3 inflammasome activation.Redox Biol. 2019 Sep;26:101254. doi: 10.1016/j.redox.2019.101254. Epub 2019 Jun 11. Redox Biol. 2019. PMID: 31229841 Free PMC article.
-
Augmenter of liver regeneration reduces mitochondria-derived ROS and NLRP3 inflammasome activation through PINK1/Parkin-mediated mitophagy in ischemia-reperfusion-induced renal tubular injury.Apoptosis. 2023 Apr;28(3-4):335-347. doi: 10.1007/s10495-022-01794-1. Epub 2022 Nov 12. Apoptosis. 2023. PMID: 36370259
-
NLRP3 Deficiency Protects Against Intermittent Hypoxia-Induced Neuroinflammation and Mitochondrial ROS by Promoting the PINK1-Parkin Pathway of Mitophagy in a Murine Model of Sleep Apnea.Front Immunol. 2021 Feb 24;12:628168. doi: 10.3389/fimmu.2021.628168. eCollection 2021. Front Immunol. 2021. PMID: 33717152 Free PMC article.
-
Tetrahydroxy stilbene glycoside alleviated inflammatory damage by mitophagy via AMPK related PINK1/Parkin signaling pathway.Biochem Pharmacol. 2020 Jul;177:113997. doi: 10.1016/j.bcp.2020.113997. Epub 2020 Apr 27. Biochem Pharmacol. 2020. PMID: 32353422
Cited by
-
Mitochondrial dynamics at the intersection of macrophage polarization and metabolism.Front Immunol. 2025 Mar 24;16:1520814. doi: 10.3389/fimmu.2025.1520814. eCollection 2025. Front Immunol. 2025. PMID: 40196123 Free PMC article. Review.
-
Research Progress on Glycolysis in the Pathogenesis of Asthma.J Asthma Allergy. 2025 Jul 26;18:1147-1160. doi: 10.2147/JAA.S528965. eCollection 2025. J Asthma Allergy. 2025. PMID: 40746369 Free PMC article. Review.
-
Combination of bone marrow mesenchymal stem cells and moxibustion restores cyclophosphamide-induced premature ovarian insufficiency by improving mitochondrial function and regulating mitophagy.Stem Cell Res Ther. 2024 Apr 8;15(1):102. doi: 10.1186/s13287-024-03709-0. Stem Cell Res Ther. 2024. PMID: 38589967 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

