HLA-DPB1 genotype variants predict DP molecule cell surface expression and DP donor specific antibody binding capacity
- PMID: 38274830
- PMCID: PMC10808447
- DOI: 10.3389/fimmu.2023.1328533
HLA-DPB1 genotype variants predict DP molecule cell surface expression and DP donor specific antibody binding capacity
Abstract
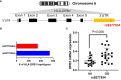
The contribution of alloresponses to mismatched HLA-DP in solid organ transplantation and hematopoietic stem cell transplantation (HCT) has been well documented. Exploring the regulatory mechanisms of DPB1 alleles has become an important question to be answered. In this study, our initial investigation focused on examining the correlation between the rs9277534G/A SNP and DPB1 mRNA expression. The result showed that there was a significant increase in DPB1 mRNA expression in B lymphoblastoid cell lines (BLCLs) with the rs9277534GG genotype compared to rs9277534AA genotype. In addition, B cells with the rs9277534GG exhibited significantly higher DP protein expression than those carrying the rs9277534AA genotype in primary B cells. Furthermore, we observed a significant upregulation of DP expression in B cells following treatment with Interleukin 13 (IL-13) compared to untreated B cells carrying rs9277534GG-linked DPB1 alleles. Fluorescence in situ hybridization (FISH) analysis of DPB1 in BLCL demonstrated significant differences in both the cytoplasmic (p=0.0003) and nuclear (p=0.0001) localization of DP mRNA expression comparing DPB1*04:01 (rs9277534AA) and DPB1*05:01 (rs9277534GG) homozygous cells. The study of the correlation between differential DPB1 expression and long non-coding RNAs (lncRNAs) showed that lnc-HLA-DPB1-13:1 is strongly associated with DP expression (r=0.85), suggesting the potential involvement of lncRNA in regulating DP expression. The correlation of DP donor specific antibody (DSA) with B cell flow crossmatch (B-FCXM) results showed a better linear correlation of DP DSA against GG and AG donor cells (R2 = 0.4243, p=0.0025 and R2 = 0.6172, p=0.0003, respectively), compared to DSA against AA donor cells (R2 = 0.0649, p=0.4244). This explained why strong DP DSA with a low expression DP leads to negative B-FCXM. In conclusion, this study provides evidence supporting the involvement of lncRNA in modulating HLA-DP expression, shedding lights on the intricate regulatory mechanisms of DP, particularly under inflammatory conditions in transplantation.
Keywords: HLA-DPB1; crossmatch; hematopoietic stem cell transplantation; long non-coding RNA; rs9277534; transplantation.
Copyright © 2024 Yin, Soe, Valenzuela, Reed and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

