An Investigation of Severe Influenza Cases in Russia during the 2022-2023 Epidemic Season and an Analysis of HA-D222G/N Polymorphism in Newly Emerged and Dominant Clade 6B.1A.5a.2a A(H1N1)pdm09 Viruses
- PMID: 38276147
- PMCID: PMC10819184
- DOI: 10.3390/pathogens13010001
An Investigation of Severe Influenza Cases in Russia during the 2022-2023 Epidemic Season and an Analysis of HA-D222G/N Polymorphism in Newly Emerged and Dominant Clade 6B.1A.5a.2a A(H1N1)pdm09 Viruses
Abstract
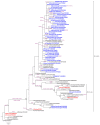
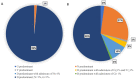
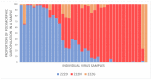
In Russia, during the COVID-19 pandemic, a decrease in influenza circulation was initially observed. Influenza circulation re-emerged with the dominance of new clades of A(H3N2) viruses in 2021-2022 and A(H1N1)pdm09 viruses in 2022-2023. In this study, we aimed to characterize influenza viruses during the 2022-2023 season in Russia, as well as investigate A(H1N1)pdm09 HA-D222G/N polymorphism associated with increased disease severity. PCR testing of 780 clinical specimens showed 72.2% of them to be positive for A(H1N1)pdm09, 2.8% for A(H3N2), and 25% for influenza B viruses. The majority of A(H1N1)pdm09 viruses analyzed belonged to the newly emerged 6B.1A.5a.2a clade. The intra-sample predominance of HA-D222G/N virus variants was observed in 29% of the specimens from A(H1N1)pdm09 fatal cases. The D222N polymorphic variant was registered more frequently than D222G. All the B/Victoria viruses analyzed belonged to the V1A.3a.2 clade. Several identified A(H3N2) viruses belonged to one of the four subclades (2a.1b, 2a.3a.1, 2a.3b, 2b) within the 3C.2a1b.2a.2 group. The majority of antigenically characterized viruses bore similarities to the corresponding 2022-2023 NH vaccine strains. Only one influenza A(H1N1)pdm09 virus showed reduced inhibition by neuraminidase inhibitors. None of the influenza viruses analyzed had genetic markers of reduced susceptibility to baloxavir.
Keywords: A(H1N1)pdm09; D222G/N polymorphism; influenza; monitoring; severe cases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Adlhoch C., Mook P., Lamb F., Ferland L., Melidou A., Amato-Gauci A.J., Pebody R., Network T.E.I.S. Decreased Influenza Activity During the COVID-19 Pandemic—United States, Australia, Chile, and South Africa, 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1305–1309. doi: 10.2807/1560-7917.ES.2021.26.11.2100221. - DOI - PMC - PubMed
-
- Adlhoch C., Mook P., Lamb F., Ferland L., Melidou A., Amato-Gauci A.J., Pebody R. European Influenza Surveillance Network.Very little influenza in the WHO European Region during the 2020/21 season, weeks 40 2020 to 8 2021. Euro Surveill. 2021;26:2100221. doi: 10.2807/1560-7917.ES.2021.26.11.2100221. - DOI - PMC - PubMed
-
- Bolton M.J., Ort J.T., McBride R., Swanson N.J., Wilson J., Awofolaju M., Furey C., Greenplate A.R., Drapeau E.M., Pekosz A., et al. Antigenic and virological properties of an H3N2 variant that continues to dominate the 2021–22 Northern Hemisphere influenza season. Cell Rep. 2022;39:110897. doi: 10.1016/j.celrep.2022.110897. - DOI - PMC - PubMed
-
- Influenza Virus Characterization: Summary Report, Europe, February 2023. WHO Regional Office for Europe and European Centre for Disease Prevention and Control; Copenhagen, Denmark: Stockholm, Sweden: 2023. [(accessed on 1 October 2023)]. Available online: https://www.ecdc.europa.eu/en/publications-data/influenza-virus-characte....
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

