Monitoring Genomic Structural Rearrangements Resulting from Gene Editing
- PMID: 38276232
- PMCID: PMC10817574
- DOI: 10.3390/jpm14010110
Monitoring Genomic Structural Rearrangements Resulting from Gene Editing
Abstract
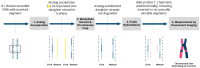
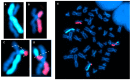
The cytogenomics-based methodology of directional genomic hybridization (dGH) enables the detection and quantification of a more comprehensive spectrum of genomic structural variants than any other approach currently available, and importantly, does so on a single-cell basis. Thus, dGH is well-suited for testing and/or validating new advancements in CRISPR-Cas9 gene editing systems. In addition to aberrations detected by traditional cytogenetic approaches, the strand specificity of dGH facilitates detection of otherwise cryptic intra-chromosomal rearrangements, specifically small inversions. As such, dGH represents a powerful, high-resolution approach for the quantitative monitoring of potentially detrimental genomic structural rearrangements resulting from exposure to agents that induce DNA double-strand breaks (DSBs), including restriction endonucleases and ionizing radiations. For intentional genome editing strategies, it is critical that any undesired effects of DSBs induced either by the editing system itself or by mis-repair with other endogenous DSBs are recognized and minimized. In this paper, we discuss the application of dGH for assessing gene editing-associated structural variants and the potential heterogeneity of such rearrangements among cells within an edited population, highlighting its relevance to personalized medicine strategies.
Keywords: DNA repair; chromosome aberrations; directional genomic hybridization; gene editing; structural variants.
Conflict of interest statement
S.M.B. and J.S.B. are cofounders and Scientific Advisory Board members of KromaTiD, Inc. E.M.C., L.K-B., H.C.S. and C.J.T. were employed by KromaTiD, Inc., at the time this work was carried out. No other interests are declared.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

