Using deep learning to quantify neuronal activation from single-cell and spatial transcriptomic data
- PMID: 38278804
- PMCID: PMC10817898
- DOI: 10.1038/s41467-023-44503-5
Using deep learning to quantify neuronal activation from single-cell and spatial transcriptomic data
Abstract
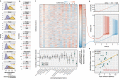
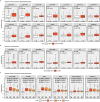
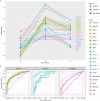
Neuronal activity-dependent transcription directs molecular processes that regulate synaptic plasticity, brain circuit development, behavioral adaptation, and long-term memory. Single cell RNA-sequencing technologies (scRNAseq) are rapidly developing and allow for the interrogation of activity-dependent transcription at cellular resolution. Here, we present NEUROeSTIMator, a deep learning model that integrates transcriptomic signals to estimate neuronal activation in a way that we demonstrate is associated with Patch-seq electrophysiological features and that is robust against differences in species, cell type, and brain region. We demonstrate this method's ability to accurately detect neuronal activity in previously published studies of single cell activity-induced gene expression. Further, we applied our model in a spatial transcriptomic study to identify unique patterns of learning-induced activity across different brain regions in male mice. Altogether, our findings establish NEUROeSTIMator as a powerful and broadly applicable tool for measuring neuronal activation, whether as a critical covariate or a primary readout of interest.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
T.A. serves on the Scientific Advisory Board of EmbarkNeuro and is a scientific advisor to Aditum Bio and Radius Health. The other authors declare no conflicting interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

