Glomerular spatial transcriptomics of IgA nephropathy according to the presence of mesangial proliferation
- PMID: 38278877
- PMCID: PMC10817934
- DOI: 10.1038/s41598-024-52581-8
Glomerular spatial transcriptomics of IgA nephropathy according to the presence of mesangial proliferation
Abstract
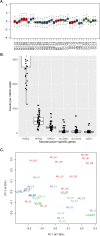
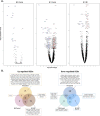
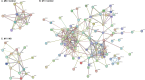
Mesangial proliferation is a diagnostic feature and a prognostic predictor of immunoglobulin A nephropathy (IgAN). We aimed to investigate the gene expression profiles of IgAN glomerulus according to the presence of mesangial proliferation. We performed spatial-specific transcriptomic profiling on kidney biopsy tissues using the GeoMx Digital Spatial Profiler. Twelve cases with three glomeruli for each case were profiled using direct pathologic classification (4 M1-IgAN, 4 M0-IgAN, and 4 donor controls). The results of enriched glom-specific genes demonstrated that M1-IgAN could be distinguished from controls (77 upregulated and 55 downregulated DEGs), while some DEGs were identified between M1-IgAN and M0-IgAN cases (24 upregulated and 8 downregulated DEGs) or between M0 and controls (1 upregulated and 16 downregulated DEGs). TCF21, an early podocyte damage marker, was the only differentially expressed gene (DEG) consistently upregulated in both M1-IgAN and M0-IgAN patients, whereas ATF3, EGR1, DUSP1, FOS, JUNB, KLF2, NR4A1, RHOB, and ZFP36 were consistently downregulated in IgAN cases. Glomeruli from M1-IgAN cases were significantly enriched for cell surface/adhesion molecules and gene expressions associated with vascular development or the extracellular matrix. Spatial transcriptomic analysis may contribute to dissecting structure-specific pathophysiology and molecular changes in IgAN.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

