Genetic architecture and biology of youth-onset type 2 diabetes
- PMID: 38278947
- PMCID: PMC10896722
- DOI: 10.1038/s42255-023-00970-0
Genetic architecture and biology of youth-onset type 2 diabetes
Abstract
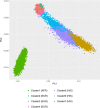
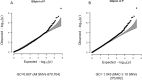
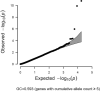
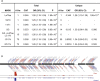
The prevalence of youth-onset type 2 diabetes (T2D) and childhood obesity has been rising steadily1, producing a growing public health concern1 that disproportionately affects minority groups2. The genetic basis of youth-onset T2D and its relationship to other forms of diabetes are unclear3. Here we report a detailed genetic characterization of youth-onset T2D by analysing exome sequences and common variant associations for 3,005 individuals with youth-onset T2D and 9,777 adult control participants matched for ancestry, including both males and females. We identify monogenic diabetes variants in 2.4% of individuals and three exome-wide significant (P < 2.6 × 10-6) gene-level associations (HNF1A, MC4R, ATXN2L). Furthermore, we report rare variant association enrichments within 25 gene sets related to obesity, monogenic diabetes and β-cell function. Many youth-onset T2D associations are shared with adult-onset T2D, but genetic risk factors of all frequencies-and rare variants in particular-are enriched within youth-onset T2D cases (5.0-fold increase in the rare variant and 3.4-fold increase in common variant genetic liability relative to adult-onset cases). The clinical presentation of participants with youth-onset T2D is influenced in part by the frequency of genetic risk factors within each individual. These findings portray youth-onset T2D as a heterogeneous disease situated on a spectrum between monogenic diabetes and adult-onset T2D.
© 2024. The Author(s).
Conflict of interest statement
A.M. receives consulting fees from Bayer, Chinook and Prokidney; research support from Alexion, Bayer, Boehringer Ingelheim and Chinook. J.C.F. has received speaking honoraria from AstraZeneca and Novo Nordisk for scientific talks over which he had full control of content; his wife has received a consulting honorarium from Novartis. The other authors declare no competing interests.
Figures








Update of
-
Insights from rare variants into the genetic architecture and biology of youth-onset type 2 diabetes.Res Sq [Preprint]. 2023 May 18:rs.3.rs-2886343. doi: 10.21203/rs.3.rs-2886343/v1. Res Sq. 2023. Update in: Nat Metab. 2024 Feb;6(2):226-237. doi: 10.1038/s42255-023-00970-0. PMID: 37292813 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
- R01 DK078616/DK/NIDDK NIH HHS/United States
- UC4 DK108173/DK/NIDDK NIH HHS/United States
- M01 RR000069/RR/NCRR NIH HHS/United States
- U01 DP000244/DP/NCCDPHP CDC HHS/United States
- P30 DK036836/DK/NIDDK NIH HHS/United States
- M01 RR014467/RR/NCRR NIH HHS/United States
- U01 DK105554/DK/NIDDK NIH HHS/United States
- U18 DP002708/DP/NCCDPHP CDC HHS/United States
- R01 DK125490/DK/NIDDK NIH HHS/United States
- U01 DK061254/DK/NIDDK NIH HHS/United States
- UL1 RR024139/RR/NCRR NIH HHS/United States
- U24 HD112205/HD/NICHD NIH HHS/United States
- UL1 RR024989/RR/NCRR NIH HHS/United States
- P30 DK057516/DK/NIDDK NIH HHS/United States
- R01 NS017950/NS/NINDS NIH HHS/United States
- UL1 RR024153/RR/NCRR NIH HHS/United States
- U18 DP006131/DP/NCCDPHP CDC HHS/United States
- U01 DP000247/DP/NCCDPHP CDC HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U18 DP002710/DP/NCCDPHP CDC HHS/United States
- K23 DK120932/DK/NIDDK NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- R03 DK138213/DK/NIDDK NIH HHS/United States
- U18 DP006134/DP/NCCDPHP CDC HHS/United States
- U18 DP006138/DP/NCCDPHP CDC HHS/United States
- UL1 RR025780/RR/NCRR NIH HHS/United States
- UL1 TR000154/TR/NCATS NIH HHS/United States
- M01 RR000036/RR/NCRR NIH HHS/United States
- U18 DP002714/DP/NCCDPHP CDC HHS/United States
- U01 DK061242/DK/NIDDK NIH HHS/United States
- U01 DP000248/DP/NCCDPHP CDC HHS/United States
- UL1 RR024992/RR/NCRR NIH HHS/United States
- UL1 TR000062/TR/NCATS NIH HHS/United States
- R01 AG008122/AG/NIA NIH HHS/United States
- U01 DK061212/DK/NIDDK NIH HHS/United States
- M01 RR001066/RR/NCRR NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- R01 AG033193/AG/NIA NIH HHS/United States
- U01 DK061230/DK/NIDDK NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- U01 HG011723/HG/NHGRI NIH HHS/United States
- U01 DK085526/DK/NIDDK NIH HHS/United States
- U18 DP006136/DP/NCCDPHP CDC HHS/United States
- U18 DP002709/DP/NCCDPHP CDC HHS/United States
- U18 DP006133/DP/NCCDPHP CDC HHS/United States
- M01 RR000043/RR/NCRR NIH HHS/United States
- UL1 TR001425/TR/NCATS NIH HHS/United States
- UM1 TR004528/TR/NCATS NIH HHS/United States
- U18 DP006139/DP/NCCDPHP CDC HHS/United States
- UL1 RR025758/RR/NCRR NIH HHS/United States
- UL1 TR000077/TR/NCATS NIH HHS/United States
- UL1 TR000423/TR/NCATS NIH HHS/United States
- M01 RR000084/RR/NCRR NIH HHS/United States
- UM1 DK078616/DK/NIDDK NIH HHS/United States
- U01 DP000250/DP/NCCDPHP CDC HHS/United States
- K24 HL157960/HL/NHLBI NIH HHS/United States
- U01 DP000246/DP/NCCDPHP CDC HHS/United States
- UL1 TR001450/TR/NCATS NIH HHS/United States
- U01 DP000254/DP/NCCDPHP CDC HHS/United States
- M01 RR000125/RR/NCRR NIH HHS/United States
- U01 DK061239/DK/NIDDK NIH HHS/United States
- HIR 10-001/HX/HSRD VA/United States
- UL1 RR024134/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

