The Dual Role of Sulforaphane-Induced Cellular Stress-A Systems Biological Study
- PMID: 38279216
- PMCID: PMC11154497
- DOI: 10.3390/ijms25021220
The Dual Role of Sulforaphane-Induced Cellular Stress-A Systems Biological Study
Abstract
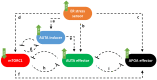
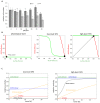
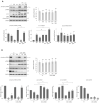
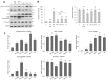
The endoplasmic reticulum (ER) plays a crucial role in cellular homeostasis. When ER stress is generated, an autophagic self-digestive process is activated to promote cell survival; however, cell death is induced in the case of excessive levels of ER stress. The aim of the present study was to investigate the effect of a natural compound called sulforaphane (SFN) upon ER stress. Our goal was to investigate how SFN-dependent autophagy activation affects different stages of ER stress induction. We approached our scientific analysis from a systems biological perspective using both theoretical and molecular biological techniques. We found that SFN induced the various cell-death mechanisms in a concentration- and time-dependent manner. The short SFN treatment at low concentrations promoted autophagy, whereas the longer treatment at higher concentrations activated cell death. We proved that SFN activated autophagy in a mTORC1-dependent manner and that the presence of ULK1 was required for its function. A low concentration of SFN pre- or co-treatment combined with short and long ER stress was able to promote cell survival via autophagy induction in each treatment, suggesting the potential medical importance of SFN in ER stress-related diseases.
Keywords: autophagy; cellular stress; feedback loops; sulforaphane; systems biology.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this article.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

