Characterization of T cell receptor repertoire in penile cancer
- PMID: 38280010
- PMCID: PMC10822009
- DOI: 10.1007/s00262-023-03615-z
Characterization of T cell receptor repertoire in penile cancer
Abstract
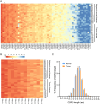
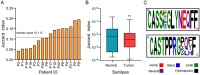
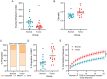
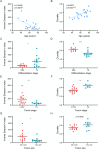
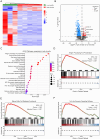
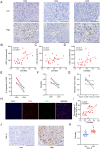
Tumor-infiltrating lymphocytes (TILs) play a key role in regulating the host immune response and shaping tumor microenvironment. It has been previously shown that T cell infiltration in penile tumors was associated with clinical outcomes. However, few studies have reported the T cell receptor (TCR) repertoire in patients with penile cancer. In the present study, we evaluated the TCR repertoires in tumor and adjacent normal tissues from 22 patients with penile squamous cell carcinoma (PSCC). Analysis of the T cell receptor beta-variable (TRBV) and joining (TRBJ) genes usage and analysis of complementarity determining region 3 (CDR3) length distribution did not show significant differences between tumor and matched normal tissues. Moreover, analysis of the median Jaccard index indicated a limited overlap of TCR repertoire between these groups. Compared with normal tissues, a significantly lower diversity and higher clonality of TCR repertoire was observed in tumor samples, which was associated with clinical characteristics. Further analysis of transcriptional profiles demonstrated that tumor samples with high clonality showed increased expression of genes associated with CD8 + T cells. In addition, we analyzed the TCR repertoire of CD4 + T cells and CD8 + T cells isolated from tumor tissues. We identified that expanded clonotypes were predominantly in the CD8 + T cell compartment, which presented with an exhausted phenotype. Overall, we comprehensively compared TCR repertoire between penile tumor and normal tissues and demonstrated the presence of distinct T cell immune microenvironments in patients with PSCC.
Keywords: High-throughput sequencing; Penile cancer; TCR clonality; TCR repertoire; Tumor microenvironment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

