Analyzing engram reactivation and long-range connectivity
- PMID: 38280198
- PMCID: PMC10840331
- DOI: 10.1016/j.xpro.2024.102840
Analyzing engram reactivation and long-range connectivity
Abstract
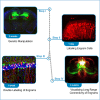
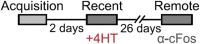
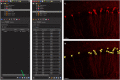
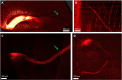
Here, we present a protocol for marking engram cells to efficiently measure reactivation levels and their projection pathways. We describe steps for genetic manipulation utilizing transgenic mice and viral infections, labeling engram cells, and a modified version of CLARITY, a tissue-clearing technique. This protocol can be adapted to various research inquiries that involve assessing the overlap of cell populations and uncovering novel long-range connectivity pathways. For complete details on the use and execution of this protocol, please refer to Refaeli et al. (2023).1.
Keywords: Behavior; Microscopy; Model Organisms; Neuroscience.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Refaeli R., Goshen I. Investigation of Spatial Interaction Between Astrocytes and Neurons in Cleared Brains. J. Vis. Exp. 2022 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

