Gut complement induced by the microbiota combats pathogens and spares commensals
- PMID: 38280374
- PMCID: PMC10922926
- DOI: 10.1016/j.cell.2023.12.036
Gut complement induced by the microbiota combats pathogens and spares commensals
Abstract
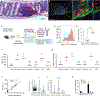
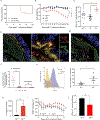
Canonically, the complement system is known for its rapid response to remove microbes in the bloodstream. However, relatively little is known about a functioning complement system on intestinal mucosal surfaces. Herein, we report the local synthesis of complement component 3 (C3) in the gut, primarily by stromal cells. C3 is expressed upon commensal colonization and is regulated by the composition of the microbiota in healthy humans and mice, leading to an individual host's specific luminal C3 levels. The absence of membrane attack complex (MAC) components in the gut ensures that C3 deposition does not result in the lysis of commensals. Pathogen infection triggers the immune system to recruit neutrophils to the infection site for pathogen clearance. Basal C3 levels directly correlate with protection against enteric infection. Our study reveals the gut complement system as an innate immune mechanism acting as a vigilant sentinel that combats pathogens and spares commensals.
Keywords: complement C3; gut complement system; innate immunity; isolated lymphoid follicles; microbiome; microbiota; stromal cells.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.R.M. is a co-founder, stakeholder, and advisor for Vizgen, Inc.
Figures





Comment in
-
Complement-ary protection for all ages.Immunity. 2024 Mar 12;57(3):411-413. doi: 10.1016/j.immuni.2024.02.010. Immunity. 2024. PMID: 38479358
References
-
- Kulkarni HS, Elvington ML, Perng Y-C, Liszewski MK, Byers DE, Farkouh C, Yusen RD, Lenschow DJ, Brody SL, and Atkinson JP (2019). Intracellular C3 Protects Human Airway Epithelial Cells from Stress-associated Cell Death. Am J Respir Cell Mol Biol 60, 144–157. 10.1165/rcmb.2017-0405OC. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

