Mitophagy curtails cytosolic mtDNA-dependent activation of cGAS/STING inflammation during aging
- PMID: 38280852
- PMCID: PMC10821893
- DOI: 10.1038/s41467-024-45044-1
Mitophagy curtails cytosolic mtDNA-dependent activation of cGAS/STING inflammation during aging
Erratum in
-
Author Correction: Mitophagy curtails cytosolic mtDNA-dependent activation of cGAS/STING inflammation during aging.Nat Commun. 2024 Feb 19;15(1):1504. doi: 10.1038/s41467-024-46010-7. Nat Commun. 2024. PMID: 38374218 Free PMC article. No abstract available.
Abstract
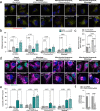
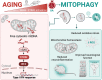
Macroautophagy decreases with age, and this change is considered a hallmark of the aging process. It remains unknown whether mitophagy, the essential selective autophagic degradation of mitochondria, also decreases with age. In our analysis of mitophagy in multiple organs in the mito-QC reporter mouse, mitophagy is either increased or unchanged in old versus young mice. Transcriptomic analysis shows marked upregulation of the type I interferon response in the retina of old mice, which correlates with increased levels of cytosolic mtDNA and activation of the cGAS/STING pathway. Crucially, these same alterations are replicated in primary human fibroblasts from elderly donors. In old mice, pharmacological induction of mitophagy with urolithin A attenuates cGAS/STING activation and ameliorates deterioration of neurological function. These findings point to mitophagy induction as a strategy to decrease age-associated inflammation and increase healthspan.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

