Chemically-defined and scalable culture system for intestinal stem cells derived from human intestinal organoids
- PMID: 38280855
- PMCID: PMC10821882
- DOI: 10.1038/s41467-024-45103-7
Chemically-defined and scalable culture system for intestinal stem cells derived from human intestinal organoids
Abstract
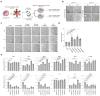
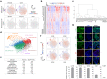
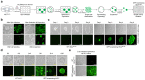
Three-dimensional human intestinal organoids (hIO) are widely used as a platform for biological and biomedical research. However, reproducibility and challenges for large-scale expansion limit their applicability. Here, we establish a human intestinal stem cell (ISC) culture method expanded under feeder-free and fully defined conditions through selective enrichment of ISC populations (ISC3D-hIO) within hIO derived from human pluripotent stem cells. The intrinsic self-organisation property of ISC3D-hIO, combined with air-liquid interface culture in a minimally defined medium, forces ISC3D-hIO to differentiate into the intestinal epithelium with cellular diversity, villus-like structure, and barrier integrity. Notably, ISC3D-hIO is an ideal cell source for gene editing to study ISC biology and transplantation for intestinal diseases. We demonstrate the intestinal epithelium differentiated from ISC3D-hIO as a model system to study severe acute respiratory syndrome coronavirus 2 viral infection. ISC3D-hIO culture technology provides a biological tool for use in regenerative medicine and disease modelling.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Xenogeneic-free culture of human intestinal stem cells on functional polymer-coated substrates for scalable, clinical-grade stem cell therapy.Nat Commun. 2024 Dec 2;15(1):10492. doi: 10.1038/s41467-024-54653-9. Nat Commun. 2024. PMID: 39622824 Free PMC article.
-
The Lumen of Human Intestinal Organoids Poses Greater Stress to Bacteria Compared to the Germ-Free Mouse Intestine: Escherichia coli Deficient in RpoS as a Colonization Probe.mSphere. 2020 Nov 11;5(6):e00777-20. doi: 10.1128/mSphere.00777-20. mSphere. 2020. PMID: 33177212 Free PMC article.
-
Effects of human induced pluripotent stem cell-derived intestinal organoids on colitis-model mice.Regen Ther. 2022 Sep 9;21:351-361. doi: 10.1016/j.reth.2022.08.004. eCollection 2022 Dec. Regen Ther. 2022. PMID: 36161099 Free PMC article.
-
Advancing Intestinal Organoid Technology Toward Regenerative Medicine.Cell Mol Gastroenterol Hepatol. 2017 Nov 2;5(1):51-60. doi: 10.1016/j.jcmgh.2017.10.006. eCollection 2018. Cell Mol Gastroenterol Hepatol. 2017. PMID: 29204508 Free PMC article. Review.
-
Engineered Synthetic Matrices for Human Intestinal Organoid Culture and Therapeutic Delivery.Adv Mater. 2024 Mar;36(9):e2307678. doi: 10.1002/adma.202307678. Epub 2023 Dec 6. Adv Mater. 2024. PMID: 37987171 Free PMC article. Review.
Cited by
-
Biomedical applications of organoids derived from the digestive system.Front Cell Dev Biol. 2025 May 30;13:1599384. doi: 10.3389/fcell.2025.1599384. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40519270 Free PMC article. Review.
-
Xenogeneic-free culture of human intestinal stem cells on functional polymer-coated substrates for scalable, clinical-grade stem cell therapy.Nat Commun. 2024 Dec 2;15(1):10492. doi: 10.1038/s41467-024-54653-9. Nat Commun. 2024. PMID: 39622824 Free PMC article.
-
Biomimetic culture substrates for modelling homeostatic intestinal epithelium in vitro.Nat Commun. 2025 May 3;16(1):4120. doi: 10.1038/s41467-025-59459-x. Nat Commun. 2025. PMID: 40316543 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

