Identification of signature gene set as highly accurate determination of metabolic dysfunction-associated steatotic liver disease progression
- PMID: 38281815
- PMCID: PMC11016492
- DOI: 10.3350/cmh.2023.0449
Identification of signature gene set as highly accurate determination of metabolic dysfunction-associated steatotic liver disease progression
Abstract
Background/aims: Metabolic dysfunction-associated steatotic liver disease (MASLD) is characterized by fat accumulation in the liver. MASLD encompasses both steatosis and MASH. Since MASH can lead to cirrhosis and liver cancer, steatosis and MASH must be distinguished during patient treatment. Here, we investigate the genomes, epigenomes, and transcriptomes of MASLD patients to identify signature gene set for more accurate tracking of MASLD progression.
Methods: Biopsy-tissue and blood samples from patients with 134 MASLD, comprising 60 steatosis and 74 MASH patients were performed omics analysis. SVM learning algorithm were used to calculate most predictive features. Linear regression was applied to find signature gene set that distinguish the stage of MASLD and to validate their application into independent cohort of MASLD.
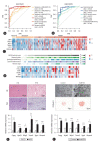
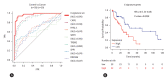
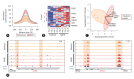
Results: After performing WGS, WES, WGBS, and total RNA-seq on 134 biopsy samples from confirmed MASLD patients, we provided 1,955 MASLD-associated features, out of 3,176 somatic variant callings, 58 DMRs, and 1,393 DEGs that track MASLD progression. Then, we used a SVM learning algorithm to analyze the data and select the most predictive features. Using linear regression, we identified a signature gene set capable of differentiating the various stages of MASLD and verified it in different independent cohorts of MASLD and a liver cancer cohort.
Conclusion: We identified a signature gene set (i.e., CAPG, HYAL3, WIPI1, TREM2, SPP1, and RNASE6) with strong potential as a panel of diagnostic genes of MASLD-associated disease.
Keywords: Biomarker; MASLD; Machine learning; Multi-omics; Signature gene set.
Conflict of interest statement
The authors have no conflicts to disclose.
Figures






Comment in
-
The gene expression signature of metabolic dysfunction- associated steatotic liver disease from a multiomics perspective.Clin Mol Hepatol. 2024 Apr;30(2):174-176. doi: 10.3350/cmh.2024.0082. Epub 2024 Feb 5. Clin Mol Hepatol. 2024. PMID: 38311818 Free PMC article. No abstract available.
References
-
- Eslam M, Sanyal AJ, George J, International Consensus Panel MAFLD: A consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158:1999–2014.e1. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

